loss
Description
L = loss(Mdl,Tbl,ResponseVarName)Mdl using the predictor data in table Tbl and
the response values in the ResponseVarName table variable. By
default, the regression loss is the mean squared error (MSE).
L = loss(___,Name=Value)
Examples
Calculate the test set mean squared error (MSE) of a regression neural network model.
Load the patients data set. Create a table from the data set. Each row corresponds to one patient, and each column corresponds to a diagnostic variable. Use the Systolic variable as the response variable, and the rest of the variables as predictors.
load patients
tbl = table(Diastolic,Height,Smoker,Weight,Systolic);Separate the data into a training set tblTrain and a test set tblTest by using a nonstratified holdout partition. The software reserves approximately 30% of the observations for the test data set and uses the rest of the observations for the training data set.
rng("default") % For reproducibility of the partition c = cvpartition(size(tbl,1),"Holdout",0.30); trainingIndices = training(c); testIndices = test(c); tblTrain = tbl(trainingIndices,:); tblTest = tbl(testIndices,:);
Train a regression neural network model using the training set. Specify the Systolic column of tblTrain as the response variable. Specify to standardize the numeric predictors, and set the iteration limit to 50.
Mdl = fitrnet(tblTrain,"Systolic", ... "Standardize",true,"IterationLimit",50);
Calculate the test set MSE. Smaller MSE values indicate better performance.
testMSE = loss(Mdl,tblTest,"Systolic")testMSE = 22.2447
Perform feature selection by comparing test set losses and predictions. Compare the test set metrics for a regression neural network model trained using all the predictors to the test set metrics for a model trained using only a subset of the predictors.
Load the sample file fisheriris.csv, which contains iris data including sepal length, sepal width, petal length, petal width, and species type. Read the file into a table.
fishertable = readtable('fisheriris.csv');Separate the data into a training set trainTbl and a test set testTbl by using a nonstratified holdout partition. The software reserves approximately 30% of the observations for the test data set and uses the rest of the observations for the training data set.
rng("default") c = cvpartition(size(fishertable,1),"Holdout",0.3); trainTbl = fishertable(training(c),:); testTbl = fishertable(test(c),:);
Train one regression neural network model using all the predictors in the training set, and train another model using all the predictors except PetalWidth. For both models, specify PetalLength as the response variable, and standardize the predictors.
allMdl = fitrnet(trainTbl,"PetalLength","Standardize",true); subsetMdl = fitrnet(trainTbl,"PetalLength ~ SepalLength + SepalWidth + Species", ... "Standardize",true);
Compare the test set mean squared error (MSE) of the two models. Smaller MSE values indicate better performance.
allMSE = loss(allMdl,testTbl)
allMSE = 0.0837
subsetMSE = loss(subsetMdl,testTbl)
subsetMSE = 0.0878
For each model, compare the test set predicted petal lengths to the true petal lengths. Plot the predicted petal lengths along the vertical axis and the true petal lengths along the horizontal axis. Points on the reference line indicate correct predictions.
tiledlayout(2,1) % Top axes ax1 = nexttile; allPredictedY = predict(allMdl,testTbl); plot(ax1,testTbl.PetalLength,allPredictedY,".") hold on plot(ax1,testTbl.PetalLength,testTbl.PetalLength) hold off xlabel(ax1,"True Petal Length") ylabel(ax1,"Predicted Petal Length") title(ax1,"All Predictors") % Bottom axes ax2 = nexttile; subsetPredictedY = predict(subsetMdl,testTbl); plot(ax2,testTbl.PetalLength,subsetPredictedY,".") hold on plot(ax2,testTbl.PetalLength,testTbl.PetalLength) hold off xlabel(ax2,"True Petal Length") ylabel(ax2,"Predicted Petal Length") title(ax2,"Subset of Predictors")
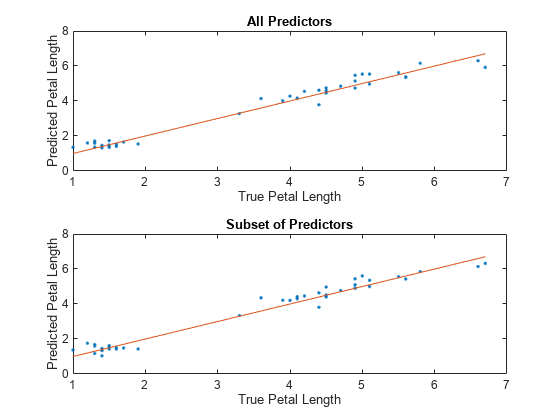
Because both models seems to perform well, with predictions scattered near the reference line, consider using the model trained using all predictors except PetalWidth.
Since R2024b
Create a regression neural network with more than one response variable.
Load the carbig data set, which contains measurements of cars made in the 1970s and early 1980s. Create a table containing the predictor variables Displacement, Horsepower, and so on, as well as the response variables Acceleration and MPG. Display the first eight rows of the table.
load carbig cars = table(Displacement,Horsepower,Model_Year, ... Origin,Weight,Acceleration,MPG); head(cars)
Displacement Horsepower Model_Year Origin Weight Acceleration MPG
____________ __________ __________ _______ ______ ____________ ___
307 130 70 USA 3504 12 18
350 165 70 USA 3693 11.5 15
318 150 70 USA 3436 11 18
304 150 70 USA 3433 12 16
302 140 70 USA 3449 10.5 17
429 198 70 USA 4341 10 15
454 220 70 USA 4354 9 14
440 215 70 USA 4312 8.5 14
Remove rows of cars where the table has missing values.
cars = rmmissing(cars);
Categorize the cars based on whether they were made in the USA.
cars.Origin = categorical(cellstr(cars.Origin)); cars.Origin = mergecats(cars.Origin,["France","Japan",... "Germany","Sweden","Italy","England"],"NotUSA");
Partition the data into training and test sets. Use approximately 85% of the observations to train a neural network model, and 15% of the observations to test the performance of the trained model on new data. Use cvpartition to partition the data.
rng("default") % For reproducibility c = cvpartition(height(cars),"Holdout",0.15); carsTrain = cars(training(c),:); carsTest = cars(test(c),:);
Train a multiresponse neural network regression model by passing the carsTrain training data to the fitrnet function. For better results, specify to standardize the predictor data.
Mdl = fitrnet(carsTrain,["Acceleration","MPG"], ... Standardize=true)
Mdl =
RegressionNeuralNetwork
PredictorNames: {'Displacement' 'Horsepower' 'Model_Year' 'Origin' 'Weight'}
ResponseName: {'Acceleration' 'MPG'}
CategoricalPredictors: 4
ResponseTransform: 'none'
NumObservations: 334
LayerSizes: 10
Activations: 'relu'
OutputLayerActivation: 'none'
Solver: 'LBFGS'
ConvergenceInfo: [1×1 struct]
TrainingHistory: [1000×7 table]
Properties, Methods
Mdl is a trained RegressionNeuralNetwork model. You can use dot notation to access the properties of Mdl. For example, you can specify Mdl.ConvergenceInfo to get more information about the model convergence.
Evaluate the performance of the regression model on the test set by computing the test mean squared error (MSE). Smaller MSE values indicate better performance. Return the loss for each response variable separately by setting the OutputType name-value argument to "per-response".
testMSE = loss(Mdl,carsTest,["Acceleration","MPG"], ... OutputType="per-response")
testMSE = 1×2
1.5341 4.8245
Predict the response values for the observations in the test set. Return the predicted response values as a table.
predictedY = predict(Mdl,carsTest,OutputType="table")predictedY=58×2 table
Acceleration MPG
____________ ______
9.3612 13.567
15.655 21.406
17.921 17.851
11.139 13.433
12.696 10.32
16.498 17.977
16.227 22.016
12.165 12.926
12.691 12.072
12.424 14.481
16.974 22.152
15.504 24.955
11.068 13.874
11.978 12.664
14.926 10.134
15.638 24.839
⋮
Input Arguments
Trained regression neural network, specified as a RegressionNeuralNetwork model object or CompactRegressionNeuralNetwork model object returned by fitrnet or
compact,
respectively.
Sample data, specified as a table. Each row of Tbl corresponds
to one observation, and each column corresponds to one predictor variable. Optionally,
Tbl can contain additional columns for the response variables and
a column for the observation weights. Tbl must contain all of the
predictors used to train Mdl. Multicolumn variables and cell arrays
other than cell arrays of character vectors are not allowed.
If
Tblcontains the response variables used to trainMdl, then you do not need to specifyResponseVarNameorY.If you trained
Mdlusing sample data contained in a table, then the input data forlossmust also be in a table.If you set
Standardize=trueinfitrnetwhen trainingMdl, then the software standardizes the numeric columns of the predictor data using the corresponding means (Mdl.Mu) and standard deviations (Mdl.Sigma).
Data Types: table
Response variable names, specified as the names of variables in
Tbl. Each response variable must be a numeric vector.
You must specify ResponseVarName as a character vector, string
array, or cell array of character vectors. For example, if Tbl stores
the response variable as Tbl.Y, then specify
ResponseVarName as "Y". Otherwise, the
software treats the Y column of Tbl as a
predictor.
Data Types: char | string | cell
Predictor data, specified as a numeric matrix. By default,
loss assumes that each row of X
corresponds to one observation, and each column corresponds to one predictor
variable.
Note
If you orient your predictor matrix so that observations correspond to columns and
specify ObservationsIn="columns", then you might experience a
significant reduction in computation time.
X and Y must have the same number of
observations.
If you set Standardize=true in fitrnet when
training Mdl, then the software standardizes the numeric columns of
the predictor data using the corresponding means (Mdl.Mu) and
standard deviations (Mdl.Sigma).
Data Types: single | double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: loss(Mdl,Tbl,"Response",Weights="W") specifies to use the
Response and W variables in the table
Tbl as the response values and observation weights,
respectively.
Loss function, specified as "mse" or a function handle.
"mse"— Weighted mean squared error.Function handle — To specify a custom loss function, use a function handle. The function must have this form:
lossval = lossfun(Y,YFit,W)
The output argument
lossvalis a floating-point scalar.You specify the function name (
lossfunIf
Mdlis a model with one response variable, thenYis a length-n numeric vector of observed responses, where n is the number of observations inTblorX. IfMdlis a model with multiple response variables, thenYis an n-by-k numeric matrix of observed responses, where k is the number of response variables.YFitis a length-n numeric vector or an n-by-k numeric matrix of corresponding predicted responses. The size ofYFitmust match the size ofY.Wis an n-by-1 numeric vector of observation weights.
Example: LossFun=@lossfun
Data Types: char | string | function_handle
Predictor data observation dimension, specified as "rows" or
"columns".
Note
If you orient your predictor matrix so that observations correspond to columns
and specify ObservationsIn="columns", then you might experience a
significant reduction in computation time. You cannot specify
ObservationsIn="columns" for predictor data in a table or for
multiresponse regression.
Data Types: char | string
Since R2024b
Type of output loss, specified as "average" or
"per-response".
| Value | Description |
|---|---|
"average" | loss averages the loss values across all
response variables and returns a scalar value. |
"per-response" | loss returns a vector, where each element
is the loss for one response variable. |
Example: OutputType="per-response"
Data Types: char | string
Since R2023b
Predicted response value to use for observations with missing predictor values,
specified as "median", "mean",
"omitted", or a numeric scalar.
| Value | Description |
|---|---|
"median" | loss uses the median of the observed
response values in the training data as the predicted response value for
observations with missing predictor values. |
"mean" | loss uses the mean of the observed
response values in the training data as the predicted response value for
observations with missing predictor values. |
"omitted" | loss excludes observations with missing
predictor values from the loss computation. |
| Numeric scalar | loss uses this value as the predicted
response value for observations with missing predictor values. |
If an observation is missing an observed response value or an observation weight, then
loss does not use the observation in the loss
computation.
Example: PredictionForMissingValue="omitted"
Data Types: single | double | char | string
Since R2024b
Flag to standardize the response data before computing the loss, specified as a
numeric or logical 0 (false) or
1 (true). If you set
StandardizeResponses to true, then the
software centers and scales each response variable by the corresponding variable mean
and standard deviation in the training data.
Specify StandardizeResponses as true when
you have multiple response variables with very different scales and
OutputType is "average". Do not standardize
the response data when you have only one response variable.
Example: StandardizeResponses=true
Data Types: single | double | logical
Observation weights, specified as a nonnegative numeric vector or the name of a
variable in Tbl. The software weights each observation in
X or Tbl with the corresponding value in
Weights. The length of Weights must equal
the number of observations in X or
Tbl.
If you specify the input data as a table Tbl, then
Weights can be the name of a variable in
Tbl that contains a numeric vector. In this case, you must
specify Weights as a character vector or string scalar. For
example, if the weights vector W is stored as
Tbl.W, then specify it as "W".
By default, Weights is ones(n,1), where
n is the number of observations in X or
Tbl.
If you supply weights, then loss computes the weighted
regression loss and normalizes weights to sum to 1.
Data Types: single | double | char | string
Output Arguments
Regression loss, returned as a numeric scalar or vector. The type of regression loss
depends on LossFun.
When Mdl is a model with one response variable,
L is a numeric scalar. When Mdl is a model
with multiple response variables, the size and interpretation of L
depend on OutputType.
Extended Capabilities
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2021aYou can create a neural network regression model with multiple response variables by
using the fitrnet function.
Regardless of the number of response variables, the function returns a
RegressionNeuralNetwork object. You can use the loss
object function to compute the regression loss on new data.
In the call to loss, you can specify to return the average loss or
the loss for each response variable by using the OutputType
name-value argument. You can also specify whether to standardize the response data before
computing the loss by using the StandardizeResponses name-value argument.
loss fully supports GPU arrays.
Starting in R2023b, when you predict or compute the loss, some regression models allow you to specify the predicted response value for observations with missing predictor values. Specify the PredictionForMissingValue name-value argument to use a numeric scalar, the training set median, or the training set mean as the predicted value. When computing the loss, you can also specify to omit observations with missing predictor values.
This table lists the object functions that support the
PredictionForMissingValue name-value argument. By default, the
functions use the training set median as the predicted response value for observations with
missing predictor values.
| Model Type | Model Objects | Object Functions |
|---|---|---|
| Gaussian process regression (GPR) model | RegressionGP, CompactRegressionGP | loss, predict, resubLoss, resubPredict |
RegressionPartitionedGP | kfoldLoss, kfoldPredict | |
| Gaussian kernel regression model | RegressionKernel | loss, predict |
RegressionPartitionedKernel | kfoldLoss, kfoldPredict | |
| Linear regression model | RegressionLinear | loss, predict |
RegressionPartitionedLinear | kfoldLoss, kfoldPredict | |
| Neural network regression model | RegressionNeuralNetwork, CompactRegressionNeuralNetwork | loss, predict, resubLoss, resubPredict |
RegressionPartitionedNeuralNetwork | kfoldLoss, kfoldPredict | |
| Support vector machine (SVM) regression model | RegressionSVM, CompactRegressionSVM | loss, predict, resubLoss, resubPredict |
RegressionPartitionedSVM | kfoldLoss, kfoldPredict |
In previous releases, the regression model loss and predict functions listed above used NaN predicted response values for observations with missing predictor values. The software omitted observations with missing predictor values from the resubstitution ("resub") and cross-validation ("kfold") computations for prediction and loss.
The loss function no longer omits an observation with a
NaN prediction when computing the weighted average regression loss. Therefore,
loss can now return NaN when the predictor data
X or the predictor variables in Tbl
contain any missing values. In most cases, if the test set observations do not contain
missing predictors, the loss function does not return
NaN.
This change improves the automatic selection of a regression model when you use
fitrauto.
Before this change, the software might select a model (expected to best predict the
responses for new data) with few non-NaN predictors.
If loss in your code returns NaN, you can update your code
to avoid this result. Remove or replace the missing values by using rmmissing or fillmissing, respectively.
The following table shows the regression models for which the
loss object function might return NaN. For more details,
see the Compatibility Considerations for each loss
function.
| Model Type | Full or Compact Model Object | loss Object Function |
|---|---|---|
| Gaussian process regression (GPR) model | RegressionGP, CompactRegressionGP | loss |
| Gaussian kernel regression model | RegressionKernel | loss |
| Linear regression model | RegressionLinear | loss |
| Neural network regression model | RegressionNeuralNetwork, CompactRegressionNeuralNetwork | loss |
| Support vector machine (SVM) regression model | RegressionSVM, CompactRegressionSVM | loss |
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)