mle
Maximum likelihood estimates
Description
phat = mle(data,Name,Value)
For example, you can specify the distribution type by using one of these name-value
arguments: Distribution, pdf,
logpdf, or nloglf.
To compute MLEs for a built-in distribution, specify the distribution type by using
Distribution. For example,'Distribution','Beta'To compute MLEs for a custom distribution, define the distribution by using
pdf,logpdf, ornloglf, and specify the initial parameter values by usingStart.
Examples
Find MLEs for a built-in distribution that you specify using the Distribution name-value argument.
Load the sample data.
load carbigThe variable MPG contains the miles per gallon for different models of cars.
Draw a histogram of the MPG data.
histogram(MPG)

The distribution is somewhat right skewed. A symmetric distribution, such as a normal distribution, might not be a good fit.
Estimate the parameters of the Burr Type XII distribution for the MPG data.
phat = mle(MPG,'Distribution','burr')
phat = 1×3
34.6447 3.7898 3.5722
The MLE for the scale parameter α is 34.6447. The estimates for the two shape parameters and of the Burr Type XII distribution are 3.7898 and 3.5722, respectively.
Generate 100 random observations from a binomial distribution with the number of trials = 20 and the probability of success = 0.75.
rng('default') % For reproducibility data = binornd(20,0.75,100,1);
Estimate the probability of success and 99% confidence limits using the simulated sample data. You must specify the number of trials (NTrials) for the binomial distribution.
[phat,pci] = mle(data,'Distribution','binomial','NTrials',20, ... 'Alpha',.01)
phat = 0.7615
pci = 2×1
0.7361
0.7856
The estimate of the probability of success is 0.7615, and the lower and upper limits of the 99% confidence interval are 0.7361 and 0.7856, respectively. This interval covers the true value used to simulate the data.
Generate sample data of size 1000 from a noncentral chi-square distribution with degrees of freedom 8 and noncentrality parameter 3.
rng default % for reproducibility x = ncx2rnd(8,3,1000,1);
Estimate the parameters of the noncentral chi-square distribution from the sample data. The Distribution name-value argument does not support the noncentral chi-square distribution. Therefore, you need to define a custom noncentral chi-square pdf using the pdf name-value argument and the ncx2pdf function. You must also specify the initial parameter values (Start name-value argument) for the custom distribution.
[phat,pci] = mle(x,'pdf',@(x,v,d)ncx2pdf(x,v,d),'Start',[1,1])
phat = 1×2
8.1052 2.6693
pci = 2×2
7.1120 1.6025
9.0983 3.7362
The estimate for the degrees of freedom is 8.1052 and the noncentrality parameter is 2.6693. The 95% confidence interval for the degrees of freedom is (7.1120,9.0983), and the interval for the noncentrality parameter is (1.6025,3.7362). The confidence intervals include the true parameter values of 8 and 3, respectively.
Load the sample data.
load('readmissiontimes.mat');The data includes ReadmissionTime, which has readmission times for 100 patients. This data is simulated.
Define a custom log pdf for a Weibull distribution with the scale parameter lambda and the shape parameter k.
custlogpdf = @(data,lambda,k) ...
log(k) - k*log(lambda) + (k-1)*log(data) - (data/lambda).^k;Estimate the parameters of the custom distribution and specify its initial parameter values (Start name-value argument).
phat = mle(ReadmissionTime,'logpdf',custlogpdf,'Start',[1,0.75])
phat = 1×2
7.5727 1.4540
The scale and shape parameters of the custom distribution are 7.5727 and 1.4540, respectively.
Load the sample data.
load('readmissiontimes.mat')The data includes ReadmissionTime, which has readmission times for 100 patients. This data is simulated.
Define a custom negative loglikelihood function for a Poisson distribution with the parameter lambda, where 1/lambda is the mean of the distribution. You must define the function to accept a logical vector of censorship information and an integer vector of data frequencies, even if you do not use these values in the custom function.
custnloglf = @(lambda,data,cens,freq) ... - length(data)*log(lambda) + sum(lambda*data,'omitnan');
Estimate the parameter of the custom distribution and specify its initial parameter value (Start name-value argument).
phat = mle(ReadmissionTime,'nloglf',custnloglf,'Start',0.05)
phat = 0.1462
Generate sample data of size 1000 from a noncentral chi-square distribution with degrees of freedom 10 and noncentrality parameter 5.
rng('default') % For reproducibility x = ncx2rnd(10,5,1000,1);
Suppose the noncentrality parameter is fixed at the value 5. Estimate the degrees of freedom of the noncentral chi-square distribution from the sample data. To do this, define a custom noncentral chi-square pdf using the pdf name-value argument.
[phat,pci] = mle(x,'pdf',@(x,v)ncx2pdf(x,v,5),'Start',1)
phat = 9.9307
pci = 2×1
9.5626
10.2989
The estimate for the noncentrality parameter is 9.9307, and the lower and upper limits of the 95% confidence interval are 9.5626 and 10.2989. The confidence interval includes the true parameter value of 10.
Add a scale parameter to the chi-square distribution for adapting to the scale of data, and fit the distribution.
Generate sample data of size 1000 from a chi-square distribution with degrees of freedom 5, and scale the data by a factor of 100.
rng default % For reproducibility x = 100*chi2rnd(5,1000,1);
Estimate the degrees of freedom and the scaling factor. To do this, define a custom chi-square probability density function using the pdf name-value argument. The density function requires a factor for data scaled by .
[phat,pci] = mle(x,'pdf',@(x,v,s)chi2pdf(x/s,v)/s,'Start',[1,200])
phat = 1×2
5.1079 99.1681
pci = 2×2
4.6862 90.1215
5.5297 108.2146
The estimate for the degrees of freedom is 5.1079 and the scale is 99.1681. The 95% confidence interval for the degrees of freedom is (4.6862,5.5279), and the interval for the scale parameter is (90.1215,108.2146). The confidence intervals include the true parameter values of 5 and 100, respectively.
Load the sample data.
load('readmissiontimes.mat');The data includes ReadmissionTime, which has readmission times for 100 patients. The column vector Censored contains the censorship information for each patient, where 1 indicates a right-censored observation, and 0 indicates that the exact readmission time is observed. This data is simulated.
Define a custom probability density function (pdf) and a cumulative distribution function (cdf) for an exponential distribution with the parameter lambda, where 1/lambda is the mean of the distribution. To fit the distribution to a censored data set, you must pass both the pdf and cdf to the mle function.
custpdf = @(data,lambda) lambda*exp(-lambda*data); custcdf = @(data,lambda) 1-exp(-lambda*data);
Estimate the parameter lambda of the custom distribution for the censored sample data. Specify the initial parameter value (Start name-value argument) for the custom distribution.
phat = mle(ReadmissionTime,'pdf',custpdf,'cdf',custcdf, ... 'Start',0.05,'Censoring',Censored)
phat = 0.1096
Generate double-censored survival data and find the MLEs for a built-in distribution of the data. Then, use the MLEs to create a probability distribution object.
Generate failure times from a Birnbaum-Saunders distribution.
rng('default') % For reproducibility failuretime = random('BirnbaumSaunders',0.3,1,[100,1]);
Assume that the study starts at time 0.1 and ends at time 0.9. The assumption implies that failure times less than 0.1 are left censored, and failure times greater than 0.9 are right censored.
Create a vector in which each element indicates the censorship status of the corresponding observation in failuretime. Use –1, 1, and 0 to indicate left-censored, right-censored, and fully observed observations, respectively.
L = 0.1; U = 0.9; left_censored = (failuretime<L); right_censored = (failuretime>U); c = right_censored - left_censored;
Find MLEs for the double-censored data. Specify the censorship information by using the Censoring name-value argument.
phat = mle(failuretime,'Distribution','BirnbaumSaunders','Censoring',c)
phat = 1×2
0.2632 1.3040
Create a probability distribution object with the MLEs by using the makedist function.
pd = makedist('BirnbaumSaunders','beta',phat(1),'gamma',phat(2))
pd =
BirnbaumSaundersDistribution
Birnbaum-Saunders distribution
beta = 0.263184
gamma = 1.304
pd is a BirnbaumSaundersDistribution object. You can use the object functions of pd to evaluate the distribution and generate random numbers. Display the supported object functions.
methods(pd)
Methods for class prob.BirnbaumSaundersDistribution: cdf gather icdf iqr mean median negloglik paramci pdf plot proflik random std truncate var
For example, compute the mean and the variance of the distribution by using the mean and var functions, respectively.
mean(pd)
ans = 0.4869
var(pd)
ans = 0.3681
Generate sample data that represents machine failure times following the Weibull distribution.
rng('default') % For reproducibility failureTimes = wblrnd(5,2,[200,1]);
Specify that observed failure times are values rounded to the nearest second.
observed = round(failureTimes);
observed is interval-censored data. An observation t in observed indicates that the event occurred after time t–0.5 and before time t+0.5.
Create a two-column matrix that includes the censorship information.
intervalTimes = [observed-0.5 observed+0.5];
The failure time must be positive. Find values smaller than eps, and change them to eps.
intervalTimes(intervalTimes < eps) = eps;
Find the MLEs for the Weibull distribution parameters by using intervalTimes.
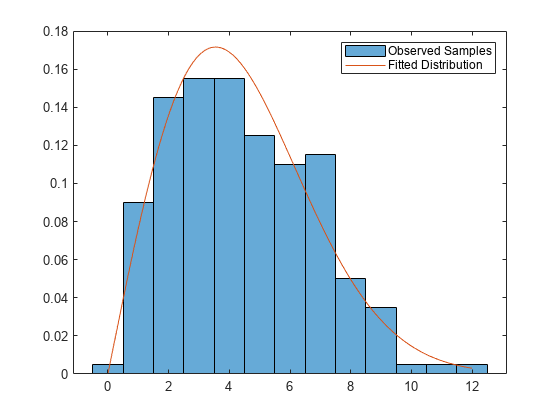
params = mle(intervalTimes,'Distribution','Weibull')
params = 1×2
5.0067 2.0049
Plot the results.
figure histogram(observed,'Normalization','pdf') hold on x = linspace(0,max(observed)); plot(x,wblpdf(x,params(1),params(2))) legend('Observed Samples','Fitted Distribution') hold off
Generate samples from a distribution with finite support, and find the MLEs with customized options for the iterative estimation process.
For a distribution with a region that has zero probability density, mle might try some parameters that have zero density, causing the function to fail to find MLEs. To avoid this problem, you can turn off the option that checks for invalid function values and specify the parameter bounds when you call the mle function.
Generate sample data of size 1000 from a Weibull distribution with the scale parameter 1 and shape parameter 1. Shift the samples by adding 10.
rng('default') % For reproducibility data = wblrnd(1,1,[1000,1]) + 10; histogram(data,'Normalization','pdf')

The histogram shows no samples smaller than 10, indicating that the distribution has zero probability in the region smaller than 10. This distribution is a three-parameter Weibull distribution, which includes a third parameter for location (see Three-Parameter Weibull Distribution).
Define a probability density function (pdf) for the three-parameter Weibull distribution.
custompdf = @(x,a,b,c) wblpdf(x-c,a,b);
Find the MLEs by using the mle function. Specify the Options name-value argument to turn off the option that checks for invalid function values. Also, specify the parameter bounds by using the LowerBound and UpperBound name-value arguments. The scale and shape parameters must be positive, and the location parameter must be smaller than the minimum of the sample data.
params = mle(data,'pdf',custompdf,'Start',[5 5 5], ... 'Options',statset('FunValCheck','off'), ... 'LowerBound',[0 0 -Inf],'UpperBound',[Inf Inf min(data)])
params = 1×3
1.0258 1.0618 10.0004
The mle function finds accurate estimates for the three parameters. For more details on specifying custom options for the iterative process, see the example Three-Parameter Weibull Distribution.
Input Arguments
Sample data and censorship information, specified as a vector of sample data or a two-column matrix of sample data and censorship information.
You can specify the censorship information for the sample data by using either the
data argument or the Censoring name-value argument.
mle ignores the Censoring argument value if
data is a two-column matrix.
Specify data as a vector or a two-column matrix depending on the
censorship types of the observations in data.
Fully observed data — Specify
dataas a vector of sample data.Data that contains fully observed, left-censored, or right-censored observations — Specify
dataas a vector of sample data, and specify theCensoringname-value argument as a vector that contains the censorship information for each observation. TheCensoringvector can contain 0, –1, and 1, which refer to fully observed, left-censored, and right-censored observations, respectively.Data that includes interval-censored observations — Specify
dataas a two-column matrix of sample data and censorship information. Each row ofdataspecifies the range of possible survival or failure times for each observation, and can have one of these values:[t,t]— Fully observed att[–Inf,t]— Left-censored att[t,Inf]— Right-censored att[t1,t2]— Interval-censored between[t1,t2], wheret1<t2
For the list of built-in distributions that support censored observations, see
Censoring.mleignoresNaNvalues indata. Additionally, anyNaNvalues in the censoring vector (Censoring) or frequency vector (Frequency) causemleto ignore the corresponding rows indata.
Data Types: single | double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: 'Censoring',Cens,'Alpha',0.01,'Options',Opt instructs
mle to estimate the parameters for the distribution of censored data
specified by the array Cens, compute the 99% confidence limits for the
parameter estimates, and use the algorithm control parameters specified by the structure
Opt.
Options to Specify Built-in Distribution
Distribution type for which to estimate parameters, specified as one of the values in this table.
Distribution Value | Distribution Type | First Parameter | Second Parameter | Third Parameter | Fourth Parameter |
|---|---|---|---|---|---|
'Bernoulli' | Bernoulli Distribution | p: probability of success for each trial | N/A | N/A | N/A |
'Beta' | Beta Distribution | a: first shape parameter | b: second shape parameter | N/A | N/A |
'Binomial' | Binomial Distribution | p: probability of success for each trial | N/A | N/A | N/A |
'BirnbaumSaunders' | Birnbaum-Saunders Distribution | β: scale parameter | γ: shape parameter | N/A | N/A |
'Burr' | Burr Type XII Distribution | α: scale parameter | c: first shape parameter | k: second shape parameter | N/A |
'Discrete Uniform' or 'unid' | Uniform Distribution (Discrete) | n: maximum observable value | N/A | N/A | N/A |
'Exponential' | Exponential Distribution | μ: mean | N/A | N/A | N/A |
'Extreme Value' or 'ev' | Extreme Value Distribution | μ: location parameter | σ: scale parameter | N/A | N/A |
'Gamma' | Gamma Distribution | a: shape parameter | b: scale parameter | N/A | N/A |
'Generalized Extreme Value' or 'gev' | Generalized Extreme Value Distribution | k: shape parameter | σ: scale parameter | μ: location parameter | N/A |
'Generalized Pareto' or 'gp' | Generalized Pareto Distribution | k: tail index (shape) parameter | σ: scale parameter | N/A | N/A |
'Geometric' | Geometric Distribution | p: probability parameter | N/A | N/A | N/A |
'Half Normal' or 'hn' | Half-Normal Distribution | σ: scale parameter | N/A | N/A | N/A |
'InverseGaussian' | Inverse Gaussian Distribution | μ: scale parameter | λ: shape parameter | N/A | N/A |
'Logistic' | Logistic Distribution | μ: mean | σ: scale parameter | N/A | N/A |
'LogLogistic' | Loglogistic Distribution | μ: mean of logarithmic values | σ: scale parameter of logarithmic values | N/A | N/A |
'LogNormal' | Lognormal Distribution | μ: mean of logarithmic values | σ: standard deviation of logarithmic values | N/A | N/A |
'Nakagami' | Nakagami Distribution | μ: shape parameter | ω: scale parameter | N/A | N/A |
'Negative Binomial' or 'nbin' | Negative Binomial Distribution | r: number of successes | p: probability of success in a single trial | N/A | N/A |
'Normal' | Normal Distribution | μ: mean | σ: standard deviation | N/A | N/A |
'Poisson' | Poisson Distribution | λ: mean | N/A | N/A | N/A |
'Rayleigh' | Rayleigh Distribution | b: scale parameter | N/A | N/A | N/A |
'Rician' | Rician Distribution | s: noncentrality parameter | σ: scale parameter | N/A | N/A |
'Stable' | Stable Distribution | α: first shape parameter | β: second shape parameter | γ: scale parameter | δ: location parameter |
'tLocationScale' | t Location-Scale Distribution | μ: location parameter | σ: scale parameter | ν: shape parameter | N/A |
'Uniform' | Uniform Distribution (Continuous) | a: lower endpoint (minimum) | b: upper endpoint (maximum) | N/A | N/A |
'Weibull' or 'wbl' | Weibull Distribution | a: scale parameter | b: shape parameter | N/A | N/A |
mle does not estimate these distribution parameters:
Number of trials for the binomial distribution. Specify the parameter by using the
NTrialsname-value argument.Location parameter of the half-normal distribution. Specify the parameter by using the
muname-value argument.Location parameter of the generalized Pareto distribution. Specify the parameter by using the
thetaname-value argument.
If the sample data is truncated or includes left-censored or interval-censored
observations, you must specify the Start name-value argument for the
Burr distribution and the stable distribution.
Example: 'Distribution','Rician'
Number of trials for the corresponding element of data for the
binomial distribution, specified as a scalar or a vector with the same number of rows as
data.
This argument is required when Distribution is
'Binomial' (binomial distribution).
Example: 'Ntrials',10
Data Types: single | double
Location (threshold) parameter for the generalized Pareto distribution, specified as a scalar.
This argument is valid only when Distribution is
'Generalized Pareto' (generalized Pareto distribution).
The default value is 0 when the sample data data includes only
nonnegative values. You must specify theta if data
includes negative values.
Example: 'theta',1
Data Types: single | double
Location parameter for the half-normal distribution, specified as a scalar.
This argument is valid only when Distribution is 'Half
Normal' (half-normal distribution).
The default value is 0 when the sample data data includes only
nonnegative values. You must specify mu if data
includes negative values.
Example: 'mu',1
Data Types: single | double
Options to Define Custom Distribution
Custom probability density function (pdf), specified as a function handle or a cell array containing a function handle and additional arguments to the function.
The custom function accepts a vector containing sample data, one or more individual distribution parameters, and any additional arguments passed by a cell array as input parameters. The function returns a vector of probability density values.
Example: 'pdf',@newpdf
Data Types: function_handle | cell
Custom cumulative distribution function (cdf), specified as a function handle or a cell array containing a function handle and additional arguments to the function.
The custom function accepts a vector containing sample data, one or more individual distribution parameters, and any additional arguments passed by a cell array as input parameters. The function returns a vector of cdf values.
To compute MLEs for censored or truncated observations, you must define both
cdf and pdf. For fully observed and untruncated
observations, mle does not use cdf. You can
specify the censorship information by using either data or
Censoring and specify the truncation bounds by using
TruncationBounds.
Example: 'cdf',@newcdf
Data Types: function_handle | cell
Custom log probability density function, specified as a function handle or a cell array containing a function handle and additional arguments to the function.
The custom function accepts a vector containing sample data, one or more individual distribution parameters, and any additional arguments passed by a cell array as input parameters. The function returns a vector of log probability values.
Example: 'logpdf',@customlogpdf
Data Types: function_handle | cell
Custom log survival function, specified as a function handle or a cell array containing a function handle and additional arguments to the function.
The custom function accepts a vector containing sample data, one or more individual distribution parameters, and any additional arguments passed by a cell array as input parameters. The function returns a vector of log survival probability values.
To compute MLEs for censored or truncated observations, you must define both
logsf and logpdf. For fully observed and
untruncated observations, mle does not use
logsf. You can specify the censorship information by using either
data or Censoring and specify the truncation
bounds by using TruncationBounds.
Example: 'logsf',@logsurvival
Data Types: function_handle | cell
Custom negative loglikelihood function, specified as a function handle or a cell array containing a function handle and additional arguments to the function.
The custom function accepts the following input arguments, in the order listed in the table.
| Input Argument of Custom Function | Description |
|---|---|
params | Vector of distribution parameter values. mle detects
the number of parameters from the number of elements in
Start. |
data | Sample data. The data value is a vector of sample data or a
two-column matrix of sample data and censorship information. |
cens | Logical vector of censorship information. nloglf must accept
cens even if you do not use the Censoring
name-value argument. In this case, you can write nloglf to ignore
cens. |
freq | Integer vector of data frequencies. nloglf must accept
freq even if you do not use the Frequency
name-value argument. In this case, you can write nloglf to ignore
freq. |
trunc | Two-element numeric vector of truncation bounds. nloglf must
accept trunc if you use the TruncationBounds
name-value argument. |
nloglf can optionally accept the additional arguments passed by a
cell array as input parameters.
nloglf returns a scalar negative loglikelihood value and, optionally,
a negative loglikelihood gradient vector (see the GradObj field in the
Options name-value argument).
Example: 'nloglf',@negloglik
Data Types: function_handle | cell
Other Options
Indicator of censored data, specified as a vector consisting of 0, –1, and 1, which
indicate fully observed, left-censored, and right-censored observations, respectively. Each
element of the Censoring value indicates the censorship status of the
corresponding observation in data. The Censoring
value must have the same size as data. The default is a vector of 0s,
indicating all observations are fully observed.
You cannot specify interval-censored observations using this argument. If the sample
data includes interval-censored observations, specify data using a
two-column matrix. mle ignores the
Censoring value if data is a two-column
matrix.
mle supports censoring for the following built-in distributions and
a custom distribution.
Distribution Value | Distribution Type |
|---|---|
'BirnbaumSaunders' | Birnbaum-Saunders |
'Burr' | Burr Type XII |
'Exponential' | Exponential |
'Extreme Value' or 'ev' | Extreme value |
'Gamma' | Gamma |
'InverseGaussian' | Inverse Gaussian |
'Logistic' | Logistic |
'LogLogistic' | Loglogistic |
'LogNormal' | Lognormal |
'Nakagami' | Nakagami |
'Normal' | Normal |
'Rician' | Rician |
'tLocationScale' | t location-scale |
'Weibull' or 'wbl' | Weibull |
For a custom distribution, you must define the distribution by using
pdf and cdf, logpdf and
logsf, or nloglf.
mle ignores any NaN values in the
censoring vector. Additionally, any NaN values in
data or the frequency vector (Frequency) cause
mle to ignore the corresponding values in the censoring
vector.
Example: 'Censoring',censored, where censored is a
vector that contains censorship information.
Data Types: logical | single | double
Truncation bounds, specified as a vector of two elements.
mle supports truncated observations for the following built-in
distributions and a custom distribution.
Distribution Value | Distribution Type |
|---|---|
'Beta' | Beta |
'BirnbaumSaunders' | Birnbaum-Saunders |
'Burr' | Burr |
'Exponential' | Exponential |
'Extreme Value' or 'ev' | Extreme value |
'Gamma' | Gamma |
'Generalized Extreme Value' or 'gev' | Generalized extreme value |
'Generalized Pareto' or 'gp' | Generalized Pareto |
'Half Normal' or 'hn' | Half-normal |
'InverseGaussian' | Inverse Gaussian |
'Logistic' | Logistic |
'LogLogistic' | Loglogistic |
'LogNormal' | Lognormal |
'Nakagami' | Nakagami |
'Normal' | Normal |
'Poisson' | Poisson |
'Rayleigh' | Rayleigh |
'Rician' | Rician |
'Stable' | Stable |
'tLocationScale' | t location-scale |
'Weibull' or 'wbl' | Weibull |
For a custom distribution, you must define the distribution by using
pdf and cdf, logpdf and
logsf, or nloglf.
Example: 'TruncationBounds',[0,10]
Data Types: single | double
Frequency of observations, specified as a vector of nonnegative integer counts that has
the same number of rows as data. The jth element of
the Frequency value gives the number of times the jth
row of data was observed. The default is a vector of 1s, indicating one
observation per row of data.
mle ignores any NaN values in this
frequency vector. Additionally, any NaN values in
data or the censoring vector (Censoring) cause
mle to ignore the corresponding values in the frequency
vector.
Example: 'Frequency',freq, where freq is a vector
that contains the observation frequencies.
Data Types: single | double
Significance level for the confidence interval pci of parameter
estimates, specified as a scalar in the range (0,1). The confidence level of
pci is 100(1–Alpha)%. The default is
0.05 for 95% confidence.
Example: 'Alpha',0.01 specifies the confidence level as
99%.
Data Types: single | double
Options for the iterative algorithm, specified as a structure returned by statset.
Use this argument to control details of the maximum likelihood optimization. This argument is valid in the following cases:
The sample data is truncated.
The sample data includes left-censored or interval-censored observations.
You fit a custom distribution.
The mle function interprets the following
statset options for optimization.
| Field Name | Description | Default Value |
|---|---|---|
GradObj | Flag indicating whether For an example of supplying a gradient to
| 'off' |
DerivStep | Relative difference, specified as a vector of the same size as
| eps^(1/3) |
FunValCheck | Flag indicating whether A poor choice for the starting point can cause
the distribution functions to return | 'on' |
TolBnd | Offset for lower and upper bounds when
| 1e-6 |
TolFun | Termination tolerance on the function value, specified as a positive scalar. | 1e-6 |
TolX | Termination tolerance for the parameters, specified as a positive scalar. | 1e-6 |
MaxFunEvals | Maximum number of function evaluations allowed, specified as a positive integer. | 400 |
MaxIter | Maximum number of iterations allowed, specified as a positive integer. | 200 |
Display | Level of display, specified as
| 'off' |
For examples of the Options name-value argument, see Find MLEs for Distribution with Finite Support and Three-Parameter Weibull Distribution.
For more details, see the options input argument of fminsearch and fmincon (Optimization Toolbox).
Example: 'Options',statset('FunValCheck','off')
Data Types: struct
Initial parameter values for the Burr distribution, stable distribution, and custom
distributions, specified as a row vector. The length of the Start value
must be the same as the number of parameters estimated by mle.
If the sample data is truncated or includes left-censored or interval-censored
observations, the Start argument is required for the Burr and stable
distributions. This argument is always required when you fit a custom distribution, that is,
when you use the pdf, logpdf, or
nloglf name-value argument. For other cases, mle
can either find initial values or compute MLEs without initial values.
Example: 0.05
Example: [100,2]
Data Types: single | double
Lower bounds for the distribution parameters, specified as a row vector of the same
length as Start.
This argument is valid in the following cases:
The sample data is truncated.
The sample data includes left-censored or interval-censored observations.
You fit a custom distribution.
Example: 'Lowerbound',0
Data Types: single | double
Upper bounds for the distribution parameters, specified as a row vector of the same
length as Start.
This argument is valid in the following cases:
The sample data is truncated.
The sample data includes left-censored or interval-censored observations.
You fit a custom distribution.
Example: 'Upperbound',1
Data Types: single | double
Optimization function used by mle to maximize the likelihood,
specified as either 'fminsearch' or 'fmincon'. The
'fmincon' option requires Optimization Toolbox™.
The sample data is truncated.
The sample data includes left-censored or interval-censored observations.
You fit a custom distribution.
Example: 'Optimfun','fmincon'
Output Arguments
Parameter estimates, returned as a row vector. For a description of parameter estimates for
the built-in distributions, see Distribution.
Confidence intervals for parameter estimates, returned as a 2-by-k matrix,
where k is the number of parameters estimated by mle.
The first and second rows of the pci show the lower and upper confidence
limits, respectively.
You can specify the significance level for the confidence interval by using the
Alpha name-value argument.
More About
mle supports left-censored, right-censored, and interval-censored observations.
Left-censored observation at time
t— The event occurred before timet, and the exact event time is unknown.Right-censored observation at time
t— The event occurred after timet, and the exact event time is unknown.Interval-censored observation within the interval
[t1,t2]— The event occurred after timet1and before timet2, and the exact event time is unknown.
Double-censored data includes both left-censored and right-censored observations.
The survival function is the probability of survival as a function of time. It is also called the survivor function.
The survival function gives the probability that the survival time of an individual exceeds a certain value. Because the cumulative distribution function F(t) is the probability that the survival time is less than or equal to a given point t in time, the survival function for a continuous distribution S(t) is the complement of the cumulative distribution function: S(t) = 1 – F(t).
Tips
When you supply custom distribution functions or use built-in distributions for left-censored, double-censored, interval-censored, or truncated observations,
mlecomputes the parameter estimates using an iterative maximization algorithm. With some models and data, a poor choice for the starting point (Start) can causemleto converge to a local optimum that is not the global maximizer, or to fail to converge entirely. Even in cases for which the loglikelihood is well behaved near the global maximum, the choice of starting point is often crucial to convergence of the algorithm. In particular, if the initial parameter values are far from the MLEs, underflow in the distribution functions can lead to infinite loglikelihoods.
Algorithms
The
mlefunction finds MLEs by minimizing the negative loglikelihood function (that is, maximizing the loglikelihood function) or by using a closed-form solution, if available. The objective function is the negative logarithm value of the product of the sample data (X) probabilities, given the distribution parameters (θ):The probability function P depends on the censorship information for each observation.
Fully observed observation — P(x|θ) = f(x), where f is the probability density function (pdf) with the parameters θ.
Left-censored observation — P(x|θ) = F(x), where F is the cumulative distribution function (cdf) with the parameters θ.
Right-censored observation — P(x|θ) = 1 – F(x).
Interval-censored observation between xL and xU — P(x|θ) = F(xU) – F(xL).
For truncated data,
mlescales the distribution functions so that all the probabilities lie in the truncation bounds [L,U].The
mlefunction computes the confidence intervalspciusing an exact method when it is available, and when the sample data is not truncated and does not include left-censored or interval-censored observations. Otherwise, the function uses the Wald method. An exact method is available for these distributions: binomial, discrete uniform, exponential, normal, lognormal, Poisson, Rayleigh, and continuous uniform.
Extended Capabilities
Usage notes and limitations:
You cannot specify the name-value argument
Distributionas'Rician'or'Stable'.If you fit a custom distribution by using the
pdfandcdf,logpdfandlogsf, ornloglfname-value arguments, the custom distribution function must support GPU arrays.
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced before R2006a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)