You have a computational task that calculates the maximum absolute eigenvalue of a
2-by-2 submatrix extracted from a large matrix. Initially, you implement this task using a
for-loop. To accelerate the computation, you convert the
for-loop into a parfor-loop, and run the
parfor-loop on a pool with six workers. When you compare the
execution times, the parfor-loop takes significantly longer than the
serial for-loop. You can use the Pool Dashboard to
investigate why the execution time for the parfor-loop is much larger
than the serial for-loop.
Open the Pool Dashboard.
To start collecting monitoring data, in the
section, select Start Monitoring. Start a parallel pool if you do
not currently have an open parallel pool.
To collect monitoring data, in the MATLAB Command Window, run this code.
n = 10000;
data = magic(n);
out = zeros(n,1);
parfor idx = 2:n
thisData = idx*data(idx-1:idx,idx-1:idx);
out(idx) = max(abs(eig(thisData)));
endVisualize the monitoring data. In the section,
select Update. The Pool Dashboard displays the
monitoring results.
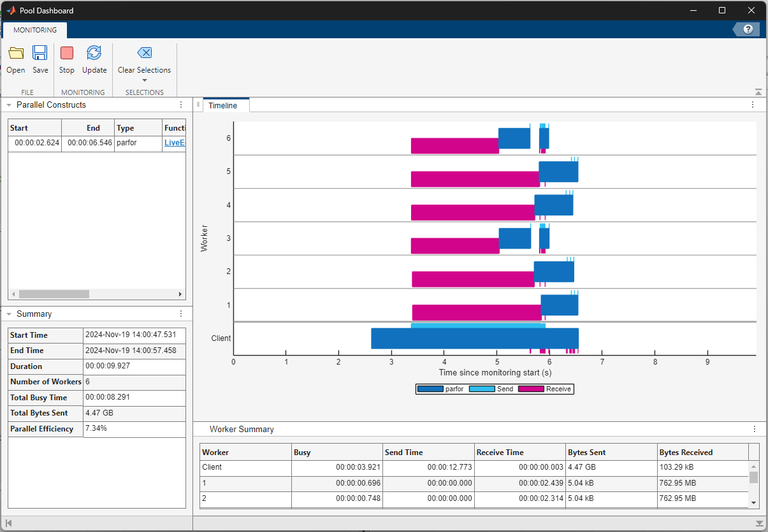
Review the monitoring data. The Timeline graph shows a visual
representation of the time the workers and client spend running the
parfor-loop and transferring data. Dark blue indicates time spent
running the parfor-loop, light blue represents time spent sending
data, and magenta represents time spent receiving data. You can observe that the workers
spend the first two to three seconds of the computation receiving data from the client.
Some workers also spend a considerable amount of time waiting to receive data from the
client.
The Worker Summary table below the
Timeline graph summarizes the information in the
Timeline graph. To view the whole table, click the three dots on
the right of the Worker Summary table and select
.
The workers spend a short time running the computations compared to transferring
data. You can observe that the client sends a total of 4.47 GB of data to the workers
and the workers each receive 762.95 MB of data while executing the
parfor-loop. The parfor-loop requires all
the workers to receive a copy of the input data, which introduces data transfer
overheads to the computation that the for-loop does not have. The
high parallel overhead dominates the computing time and this indicates the
for-loop does not benefit from conversion into a
parfor-loop.
However, if you need to run a parfor-loop multiple times using
the same set of data, you can optimize the parfor-loop by
transferring the input data to the workers only once using a Constant
object. This is a one off cost, and the workers have access to the data until you clear
the Constant object.
To understand how using a Constant object optimizes the
parfor-loop, run this code in the Command
Window.
n = 10000;
data = magic(n);
out = zeros(n,1);
C = parallel.pool.Constant(data);
parfor idx = 1:10
c = C.Value;
end
parfor idx = 2:n
thisData = idx.*C.Value(idx-1:idx,idx-1:idx);
out(idx) = max(abs(eig(thisData)));
endVisualize the monitoring data. On the
tab, in the
section, select
Update.
The Pool Dashboard updates the displayed monitoring results.
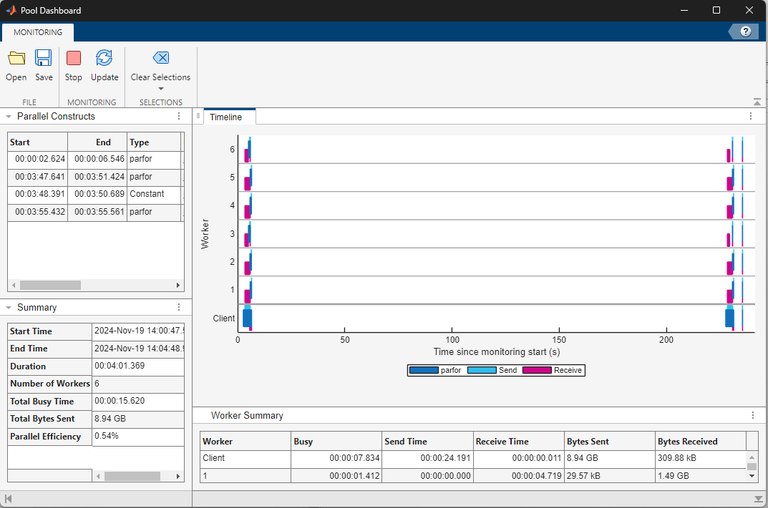
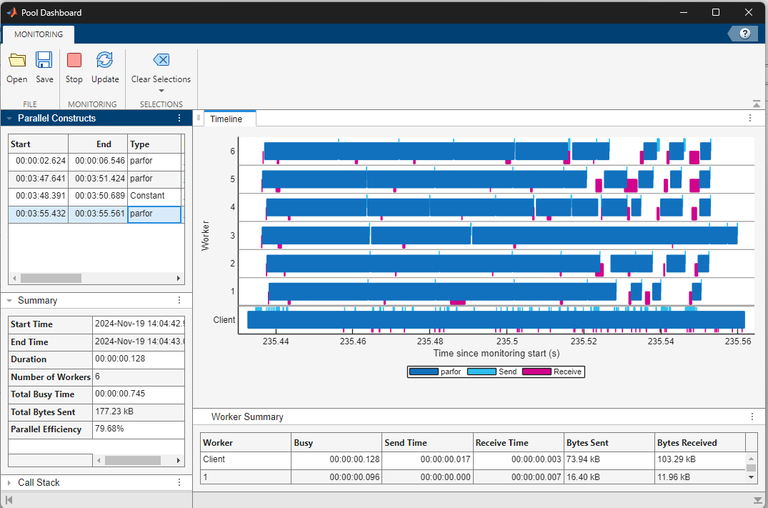
To view the monitoring data for only the parfor-loop that uses
the Constant object, in the Parallel Constructs panel, select the last
row in the table. When you use the Constant object to transfer data to
the workers before you run the parfor-loop, the workers only spend
time running the computations.
To stop collecting monitoring data, in the
section, select Stop.