MultinomialRegression
Description
MultinomialRegression is a fitted multinomial regression model
object. A multinomial regression model describes the relationship between predictors and a
response that has a finite set of values.
Use the properties of a MultinomialRegression object to investigate a
fitted multinomial regression model. The object properties include information about
coefficient estimates, summary statistics, and the data used to fit the model. Use the object
functions to predict responses, and to evaluate and visualize the multinomial regression
model.
Creation
Create a MultinomialRegression model object with specified parameter
values by using fitmnr.
Properties
Coefficient Estimates
This property is read-only.
Names of the response variable categories used to fit the multinomial regression
model, specified as a k-by-1 categorical array, character array,
logical vector, numeric vector, or cell array of character vectors.
k is the number of response categories.
ClassNames has the same data type as the response category
labels. Note that the software treats string arrays as cell arrays of character
vectors. The ClassNames property is set by the fitmnr
input argument Y or Tbl when you create the
model object.
Data Types: single | double | logical | char | cell | categorical
This property is read-only.
Covariance matrix for model coefficients, specified as a (p+1)-by-(p+1) matrix of numeric values. p is the number of predictor variables.
For details, see Coefficient Standard Errors and Confidence Intervals.
Data Types: single | double
This property is read-only.
Coefficient names, specified as a cell array of character vectors, each containing
the name of the corresponding coefficient. Each coefficient name is the name of a
response category appended to the name of a predictor or intercept. This property is
set by the fitmnr
input argument Tbl or name-value argument
PredictorNames when you create the model object.
Data Types: cell
This property is read-only.
Coefficient values, specified as a table that contains one row for each coefficient and these columns:
Value— Estimated coefficient valueSE— Standard error of the estimatetStat— t-statistic for a two-sided test with the null hypothesis that the coefficient is zeropValue— p-value for the t-statistic
Use coefTest or
testDeviance to perform other tests on the coefficients. Use coefCI to
find the confidence intervals of the coefficient estimates.
Data Types: table
This property is read-only.
Indicator for an interaction between response categories and coefficients,
specified as a numeric or logical 1 (true) or
0 (false). This property is set by the
fitmnr
name-value argument IncludeClassInteractions when you create the
model object.
Data Types: logical
This property is read-only.
Link function to use for ordinal and hierarchical models, specified as
'logit', 'probit',
'comploglog', or 'loglog'. For nominal models,
Link is always 'logit'. This property is set
by the fitmnr
name-value argument Link when you create the model object.
Data Types: char
This property is read-only.
Type of model, specified as 'nominal',
'ordinal', or 'hierarchical'. This property is
set by the fitmnr
name-value argument ModelType when you create the model
object.
Data Types: char
This property is read-only.
Number of model coefficients, specified as a positive integer.
Data Types: double
Summary Statistics
This property is read-only.
Deviance of the fit, specified as a numeric value. The deviance is useful for comparing two models when one model is a special case of the other model. The difference between the deviance of the two models has a chi-square distribution with degrees of freedom equal to the difference in the number of estimated parameters between the two models. For more information, see Deviance.
Data Types: single | double
This property is read-only.
Degrees of freedom for the error (residuals), specified as a positive integer. For
nominal and ordinal models, DFE is given by
where n is the number of observations,
k is the number of response categories, and N
is the number of model coefficients. For hierarchical models, DFE
is given by
when IncludeClassInteractions is false. When
IncludeClassInteractions is true,
DFE for a hierarchical model is given by
where ni is the number of observations corresponding to the ith response category and above.
Data Types: double
This property is read-only.
Variance, specified as a numeric scalar. If you set the fitmnr
EstimateDispersion name-value argument to true
when you create the model object, the function estimates the standard error as the
Dispersion value. Otherwise, fitmnr
assigns the default theoretical value of 1 to Dispersion.
Data Types: single | double
This property is read-only.
Indicator for whether dispersion is estimated, specified as a logical
false or true. This property is set by the
fitmnr
EstimateDispersion name-value argument when you create the model
object.
Data Types: single | double | logical
This property is read-only.
Fitted (predicted) response values based on the input data, specified as an
n-by-1 categorical array, character array, logical vector,
numeric vector, or cell array of character vectors. n is the number
of observations in the input data. Fitted has the same data type
as the response category labels. Note that the software treats string arrays as cell
arrays of character vectors. Use predict to
compute the predictions for other predictor values, or to compute the confidence
bounds on Fitted.
Data Types: single | double | logical | char | cell | categorical
This property is read-only.
Loglikelihood of the fitted model, specified as a numeric value, based on the
assumption that each response value follows a multinomial distribution. When you
create the model object, fitmnr
calculates the loglikelihood of the model by taking the sum of the log probabilities
for the response data.
Data Types: single | double
This property is read-only.
Criterion for model comparison, specified as a structure with these fields:
AIC— Akaike information criterion.AIC = –2*lnL + 2*m, wherelnLis the loglikelihood andmis the number of estimated parameters.AICc— Akaike information criterion corrected for the sample size.AICc = AIC + (2*m*(m + 1))/(n – m – 1), wherenis the number of observations.BIC— Bayesian information criterion.BIC = –2*lnL + m*ln(n).CAIC— Consistent Akaike information criterion.CAIC = –2*lnL + m*(ln(n) + 1).
Information criteria are model selection tools you can use to compare multiple models that are fit to the same data. These criteria are likelihood-based measures of model fit that include a penalty for complexity (specifically, the number of parameters). Different information criteria are distinguished by the form of the penalty.
When you compare multiple models, the model with the lowest information criterion value is the best-fitting model. The best-fitting model can vary depending on the criterion used for model comparison.
Data Types: struct
This property is read-only.
Residuals for the fitted model, specified as a table in which each variable contains one row for each observation and one column for each response class.
| Column | Description |
|---|---|
Raw | Raw residuals. Observed minus fitted values,
|
Pearson | Raw residuals divided by the root mean squared error (RMSE) |
Deviance | Deviance residuals given by the formula |
Rows not used in the fit because of missing values contain NaN
values. To inspect missing values, see ObservationInfo.
Use plotResiduals to create a plot of the residuals. For
details, see Residuals.
Data Types: table
This property is read-only.
Pseudo R-squared values for the fitted model, specified as a structure. Each field
of Rsquared contains a pseudo R-squared value calculated with a
different formula [1].
| Field | Description |
|---|---|
'Ordinary' | The ordinary pseudo R-squared value is where is the loglikelihood of the fitted model and is the loglikelihood of a model with no predictors. |
'Adjusted' | The adjusted pseudo R-squared value is where K is the number of model coefficients in . |
Data Types: struct
Input Data
This property is read-only.
Regression model, specified as a LinearFormula object. This
property is set by the fitmnr
input argument Formula when you create the model object.
This property is read-only.
Number of observations used by the fitting algorithm to fit the model, specified
as a positive integer. NumObservations is the number of
observations supplied in the original table or matrix, minus any rows with missing
values.
Data Types: double
This property is read-only.
Number of predictor variables used by the fitting algorithm to fit the model, specified as a positive integer.
Data Types: double
This property is read-only.
Number of variables in the input data, specified as a positive integer.
NumVariables includes any variables that are not used as
predictors or as the response to fit the model.
Data Types: double
This property is read-only.
Observation information, specified as an n-by-3 table containing the following columns, where n is the number of observations.
| Column | Description |
|---|---|
Weights | Observation weights, specified as a numeric value. The default value is
1. |
Missing | Indicator of missing observations, specified as a logical value. The
value is true if the observation is missing. |
Subset | Indicator of whether fitmnr uses the observation, specified as a logical value. The
value is true if the observation is not missing, meaning
fitmnr uses the observation. |
Data Types: table
This property is read-only.
Observation names, specified as a cell array of character vectors containing the names of the observations used in the fit.
If the fit is based on a table or dataset containing observation names, the
ObservationNamesproperty contains those names.Otherwise,
ObservationNamesis an empty cell array.
This property is set by the fitmnr
input argument Tbl when you create the model object and assign
row names to Tbl.
Data Types: cell
This property is read-only.
Names of the predictors used to fit the model, specified as a cell array of
character vectors. This property is set by one of the following fitmnr
arguments when you create the model object:
Tblinput argumentXinput argument together with thePredictorNamesname-value argument
Data Types: cell
This property is read-only.
Response variable name, specified as a character vector. This property is set by
one of the following fitmnr
arguments when you create the model object:
ResponseNamename-value argumentTblinput argument together with theResponseVarNameinput argumentTblinput argument together with theFormulainput argument
Data Types: char
This property is read-only.
Information about the variables contained in the Variables property, specified as a table with one row for each variable and the following columns.
| Column | Description |
|---|---|
Class | Variable class, specified as a cell array of character vectors, such as
'double' and 'categorical' |
Range | Variable range, specified as a cell array of vectors
|
InModel | Indicator of which variables are in the fitted model, specified as a
logical vector. The value is true if the model includes the
variable. |
IsCategorical | Indicator of categorical variables, specified as a logical vector. The
value is true if the variable is categorical. |
VariableInfo also includes any variables that are not used as
predictors or as the response to fit the model.
Data Types: table
This property is read-only.
Names of the variables, specified as a cell array of character vectors. Elements
of this property are set by one of the following fitmnr
arguments when you create the model object:
The
Tblinput argument specifies the names of the predictor variables, response, and unused variables.The
PredictorNamesname-value argument specifies the names of the predictor variables.The
ResponseVarNamename-value argument specifies the name of the response variable.
VariableNames also includes any variables that are not used as
predictors or as the response to fit the model.
Data Types: cell
This property is read-only.
Input data, specified as a table. Variables contains both
predictor and response values. Elements of this property are set by one of the
following fitmnr
arguments when you create the model object:
If you specify
X, thenVariablescontains all variables in the columns ofX.If you specify
Tbl, thenVariablescontains all variables inTbl, including variables not used as predictor or response data to fit the model.If you specify
Y, thenVariablesalso contains the response data inY.
Data Types: table
Object Functions
coefCI | Confidence intervals for coefficient estimates of multinomial regression model |
coefTest | Linear hypothesis test on multinomial regression model coefficients |
feval | Predict responses of multinomial regression model using one input for each predictor |
partialDependence | Compute partial dependence |
plotPartialDependence | Create partial dependence plot (PDP) and individual conditional expectation (ICE) plots |
plotResiduals | Plot residuals of multinomial regression model |
plotSlice | Plot of slices through fitted multinomial regression surface |
predict | Predict responses of multinomial regression model |
random | Generate random responses from fitted multinomial regression model |
testDeviance | Deviance test for multinomial regression model |
Examples
Load the fisheriris sample data set.
load fisheririsThe column vector species contains iris flowers of three different species: setosa, versicolor, virginica. The matrix meas contains four types of measurements for the flower: the length and width of sepals and petals in centimeters.
Fit a multinomial regression model to predict the iris flower species using the measurements. Display the results of the fit using the Coefficients property of the fitted model.
MnrModel = fitmnr(meas,species); MnrModel.Coefficients
ans=10×4 table
Value SE tStat pValue
_______ ______ _______ __________
(Intercept_setosa) 1848.8 12.404 149.05 0
x1_setosa 617.39 3.5783 172.54 0
x2_setosa -521.06 3.176 -164.06 0
x3_setosa -472.64 3.5403 -133.5 0
x4_setosa -2530.7 7.1203 -355.42 0
(Intercept_versicolor) 42.638 5.2719 8.0878 6.0776e-16
x1_versicolor 2.4652 1.1228 2.1956 0.028124
x2_versicolor 6.6809 1.4789 4.5176 6.2559e-06
x3_versicolor -9.4294 1.2934 -7.2906 3.0859e-13
x4_versicolor -18.286 2.0967 -8.7214 2.7475e-18
MnrModel is a multinomial regression model object that contains the results of fitting a nominal multinomial regression model to the data. The Coefficients property contains coefficient statistics for each predictor in meas. The small p-values in the column pValue indicate that all coefficients are statistically significant at the 95% confidence level. fitmnr sorts the categories in species in order of their first appearance. The last category is the default reference category.
To display the sorted names of the response variable categories, use the ClassNames property of MnrModel.
MnrModel.ClassNames
ans = 3×1 cell
{'setosa' }
{'versicolor'}
{'virginica' }
The output shows that the last category, 'virginica', is the reference category by default.
To get 95% confidence intervals for the fitted coefficient estimates, call the object function coefCI.
coefCI(MnrModel)
ans = 10×2
103 ×
1.8243 1.8732
0.6104 0.6244
-0.5273 -0.5148
-0.4796 -0.4657
-2.5447 -2.5167
0.0323 0.0530
0.0003 0.0047
0.0038 0.0096
-0.0120 -0.0069
-0.0224 -0.0142
The output shows 95% confidence intervals for the 10 coefficients in the Value column of the Coefficients table. None of the confidence intervals cross zero, confirming that all coefficients affect the log odds at the 95% confidence level.
Load the fisheriris sample data set.
load fisheririsThe column vector species contains three iris flowers species: setosa, versicolor, and virginica. The matrix meas contains four types of measurements for the flower: the length and width of sepals and petals in centimeters.
Divide the species and measurement data into training and test data by using the cvpartition function. Get the indices of the training data rows by using the training function.
n = length(species);
partition = cvpartition(n,'Holdout',0.05);
idx_train = training(partition);Create training data by using the indices of the training data rows to create a matrix of measurements and a vector of species labels.
meastrain = meas(idx_train,:); speciestrain = species(idx_train,:);
Fit a multinomial regression model using the training data.
mdl = fitmnr(meastrain,speciestrain)
mdl =
Multinomial regression with nominal responses
Value SE tStat pValue
_______ ______ ________ __________
(Intercept_setosa) 86.305 12.541 6.8817 5.9158e-12
x1_setosa -1.0728 3.5795 -0.29971 0.7644
x2_setosa 23.846 3.1238 7.6336 2.2835e-14
x3_setosa -27.289 3.5009 -7.795 6.4409e-15
x4_setosa -59.58 7.0214 -8.4855 2.1472e-17
(Intercept_versicolor) 42.637 5.2214 8.1659 3.1906e-16
x1_versicolor 2.4652 1.1263 2.1887 0.028619
x2_versicolor 6.6808 1.474 4.5325 5.829e-06
x3_versicolor -9.4292 1.2946 -7.2837 3.248e-13
x4_versicolor -18.286 2.0833 -8.7775 1.671e-18
143 observations, 276 error degrees of freedom
Dispersion: 1
Chi^2-statistic vs. constant model: 302.0378, p-value = 1.5168e-60
mdl is a multinomial regression model object that contains the results of fitting a nominal multinomial regression model to the data. The table output shows coefficient statistics for each predictor in meas. By default, fitmnr uses virginica as the reference category.
Get the indices of the test data rows by using the test function. Create test data by using the indices of the test data rows to create a matrix of measurements and a vector of species labels.
idx_test = test(partition); meastest = meas(idx_test,:); speciestest = species(idx_test,:);
Predict the iris species for the measurements in meastest.
speciespredict = predict(mdl,meastest)
speciespredict = 7×1 cell
{'setosa' }
{'setosa' }
{'setosa' }
{'setosa' }
{'setosa' }
{'versicolor'}
{'versicolor'}
Compare the predictions in speciespredict with the category names in speciestest.
speciestest
speciestest = 7×1 cell
{'setosa' }
{'setosa' }
{'setosa' }
{'setosa' }
{'setosa' }
{'versicolor'}
{'versicolor'}
The output shows that the model accurately predicts the iris species for the measurements in meastest.
Load the carbig sample data set.
load carbig;The vectors Acceleration and Displacement contain data for car acceleration and displacement, respectively. The vector Cylinders contains data for the number of cylinders in each car engine.
Fit an ordinal multinomial regression model using Acceleration and Displacement as predictor variables and Cylinders as the response variable.
MnrModel = fitmnr([Acceleration,Displacement],Cylinders,Model="ordinal",... PredictorNames=["Acceleration" "Displacement"])
MnrModel =
Multinomial regression with ordinal responses
Value SE tStat pValue
_________ ________ _______ __________
(Intercept_3) 11.949 3.1817 3.7555 0.00017299
(Intercept_4) 27.08 4.9481 5.4727 4.4321e-08
(Intercept_5) 27.528 4.9738 5.5346 3.1195e-08
(Intercept_6) 45.346 7.8292 5.7919 6.9593e-09
Acceleration -0.063533 0.1041 -0.6103 0.54167
Displacement -0.16731 0.027885 -6 1.9726e-09
406 observations, 1618 error degrees of freedom
Dispersion: 1
Chi^2-statistic vs. constant model: 786.5846, p-value = 1.5679e-171
MnrModel is a multinomial regression model object that contains the results of fitting an ordinal multinomial regression model to the data. The table output shows coefficient statistics for each predictor variable. The p-values in the column pValue indicate that there is not enough evidence to conclude that the coefficient for the Acceleration term is statistically significant. However, enough evidence exists to conclude that Displacement has a statistically significant effect at the 99% confidence level.
Display the possible quantities for car engine cylinders using the ClassNames property.
MnrModel.ClassNames
ans = 5×1
3
4
5
6
8
The last category in the output is the default reference category. The output shows that the reference category corresponds to cars with eight-cylinder engines.
Use plotSlice to plot stacked histograms of the probabilities of a car having each number of cylinders as the value of the predictor variable Displacement changes. By default, plotSlice fixes the value of Acceleration at its training data mean.
plotSlice(MnrModel,"stackedhist",PredictorToVary="Displacement") hold on lgd = legend; title(lgd, "Number of cylinders");
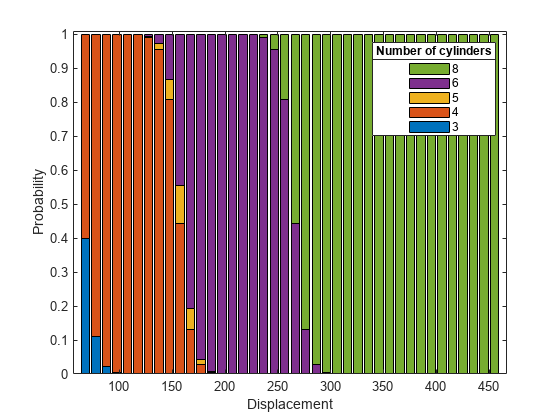
The plot shows that the probability of a car having more cylinders increases as the car displacement increases, which is consistent with the small p-value for the Displacement model term.
Load the carbig sample data set.
load carbig;The vectors Acceleration and Displacement contain data for car acceleration and displacement, respectively. The vector Cylinders contains data for the number of cylinders in each car engine.
Fit an ordinal multinomial regression model using Acceleration and Displacement as predictor variables and Cylinders as the response variable.
MnrModel = fitmnr([Acceleration,Displacement],Cylinders,Model="ordinal",... PredictorNames=["Acceleration" "Displacement"])
MnrModel =
Multinomial regression with ordinal responses
Value SE tStat pValue
_________ ________ _______ __________
(Intercept_3) 11.949 3.1817 3.7555 0.00017299
(Intercept_4) 27.08 4.9481 5.4727 4.4321e-08
(Intercept_5) 27.528 4.9738 5.5346 3.1195e-08
(Intercept_6) 45.346 7.8292 5.7919 6.9593e-09
Acceleration -0.063533 0.1041 -0.6103 0.54167
Displacement -0.16731 0.027885 -6 1.9726e-09
406 observations, 1618 error degrees of freedom
Dispersion: 1
Chi^2-statistic vs. constant model: 786.5846, p-value = 1.5679e-171
MnrModel is a multinomial regression model object that contains the results of fitting an ordinal multinomial regression model to the data. The table output shows coefficient statistics for each of the predictor variable. The p-values in the column pValue indicate that there is not enough evidence to conclude that the coefficient for the Acceleration term is statistically significant. However, enough evidence exists to conclude that Displacement has a statistically significant effect at the 99% confidence level.
Display the possible quantities for car engine cylinders using the ClassNames property.
MnrModel.ClassNames
ans = 5×1
3
4
5
6
8
The reference category corresponds to cars with eight-cylinder engines.
Plot the partial dependence of the reference category probability on the Displacement predictor by using the plotPartialDependence object function.
plotPartialDependence(MnrModel,2,8)
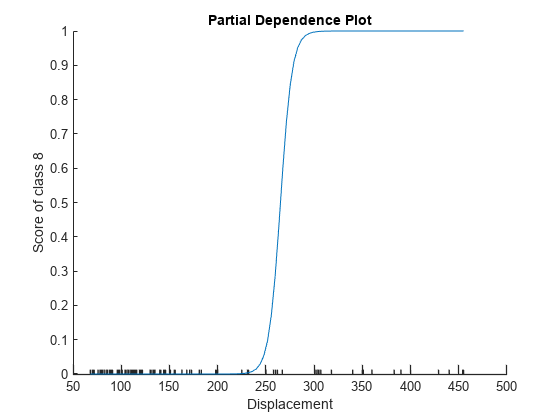
The plot shows that the probability of a car being in the reference category increases sharply when the value of Displacement reaches approximately 250.
More About
Deviance is a generalization of the residual sum of squares. It measures the goodness of fit compared to a saturated model.
The deviance of a model M1 is twice the difference between the loglikelihood of the model M1 and the saturated model Ms. A saturated model is a model with the maximum number of parameters that you can estimate.
For example, if you have n observations with potentially different response values yi, i = 1, 2, ..., n, then you can define a saturated model (with n parameters) that perfectly predicts the responses. Let L(b,y) denote the maximum value of the likelihood function for a model with the parameters b. Then the deviance of the model M1 is
where b1 and bs contain the estimated parameters for the model M1 and the saturated model, respectively. The deviance has a chi-square distribution with n – p degrees of freedom, where n is the number of parameters in the saturated model and p is the number of parameters in the model M1.
Assume you have two different multinomial regression models M1 and M2, and M1 has a subset of the terms in M2. You can evaluate the fit of the models by comparing the deviances D1 and D2 of the two models. The difference of the deviances is
Asymptotically, the difference D has a chi-square distribution with
degrees of freedom v equal to the difference in the number of
parameters estimated in M1 and
M2. You can obtain the
p-value for this test by using
1 – chi2cdf(D,v,"upper").
Typically, you examine D using a model M2 with a constant term and no predictors. Therefore, D has a chi-square distribution with p – 1 degrees of freedom. If the dispersion is estimated, the difference divided by the estimated dispersion has an F distribution with p – 1 numerator degrees of freedom and n – p denominator degrees of freedom.
References
[1] Allison, P. D. "Measures of Fit for Logistic Regression." Statistical Horizons LLC and the University of Pennsylvania, 2014.
[2] McCullagh, P., and J. A. Nelder. Generalized Linear Models. New York: Chapman & Hall, 1990.
[3] Long, J. S. Regression Models for Categorical and Limited Dependent Variables. Sage Publications, 1997.
[4] Dobson, A. J., and A. G. Barnett. An Introduction to Generalized Linear Models. Chapman and Hall/CRC. Taylor & Francis Group, 2008.
Version History
Introduced in R2023a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)