medfilt1
1-D median filtering
Syntax
Description
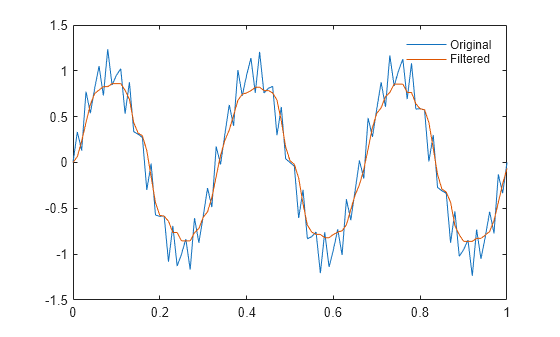
Examples
Input Arguments
Output Arguments
Tips
If you have a license for Image Processing Toolbox™ software,
you can use the medfilt2 (Image Processing Toolbox) function
to perform two-dimensional median filtering.
References
[1] Pratt, William K. Digital Image Processing. 4th Ed. Hoboken, NJ: John Wiley & Sons, 2007.
Extended Capabilities
Version History
Introduced before R2006a
See Also
filter | hampel | median | movmedian | sgolayfilt