imsegsam
Perform automatic full image segmentation using Segment Anything Model (SAM)
Since R2024b
Description
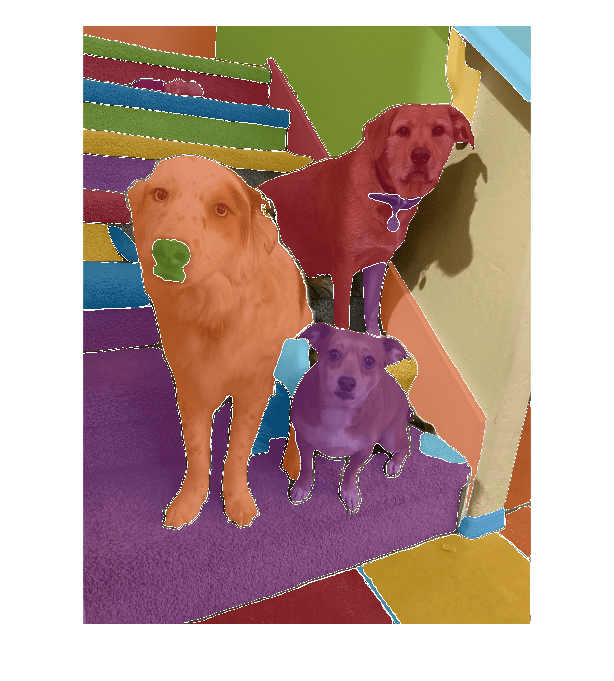
Use the imsegsam function to automatically segment the entire
image or all of the objects inside an ROI using the Segment Anything Model (SAM). The SAM
samples a regular grid of points on an image and returns a set of predicted masks for each
point, which enables the model to produce multiple masks for each object and its subregions.
You can customize various segmentation settings based on your application, such as the ROI in
which to segment objects, the size range of objects which to segment, and the confidence score
threshold with which to filter mask predictions.
Note
This functionality requires Deep Learning Toolbox™, Computer Vision Toolbox™, and the Image Processing Toolbox™ Model for Segment Anything Model. You can install the Image Processing Toolbox Model for Segment Anything Model from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
[
specifies options using one or more name-value arguments. For example,
masks,scores] = imsegsam(I,Name=Value)PointGridSize=[64 64] specifies the number of grid points that the
imsegsam function samples along the x- and
y- directions of the input image as 64
each.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Kirillov, Alexander, Eric Mintun, Nikhila Ravi, Hanzi Mao, Chloe Rolland, Laura Gustafson, Tete Xiao, et al. "Segment Anything," April 5, 2023. https://doi.org/10.48550/arXiv.2304.02643.
Version History
Introduced in R2024b
See Also
Functions
segmentAnythingModel|labelmatrix|labeloverlay|insertObjectMask(Computer Vision Toolbox)
Apps
- Image Segmenter | Image Labeler (Computer Vision Toolbox)