isanomaly
Find anomalies in data using one-class support vector machine (SVM) for incremental learning
Since R2023b
Syntax
Description
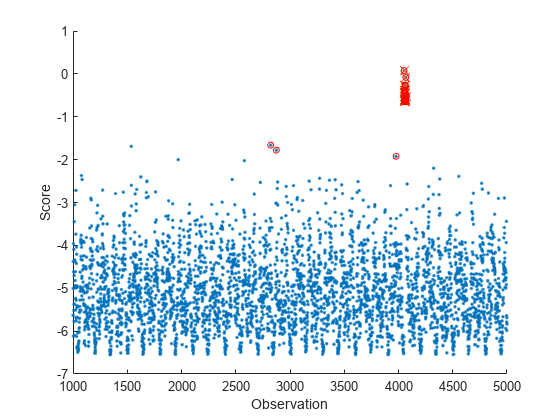
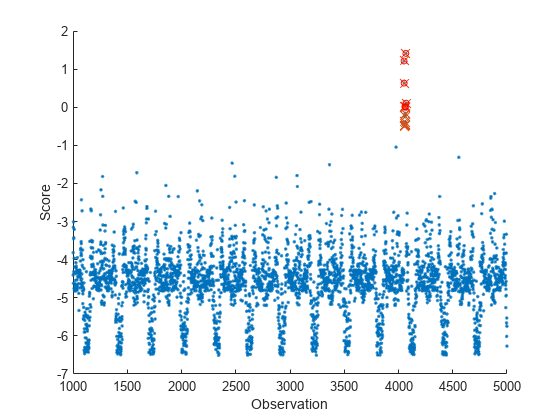
tf = isanomaly(Mdl,Tbl)Tbl using the incrementalOneClassSVM object Mdl and returns the logical
array tf, whose elements are true when an anomaly is
detected in the corresponding row of Tbl. You must use this syntax if
you create Mdl by passing a table to incrementalOneClassSVM or the incrementalLearner function of OneClassSVM.
tf = isanomaly(___,ScoreThreshold=scoreThreshold)isanomaly detects observations with scores
above scoreThreshold as anomalies.
Examples
Input Arguments
Output Arguments
References
[1] Guha, Sudipto, N. Mishra, G. Roy, and O. Schrijvers. "Robust Random Cut Forest Based Anomaly Detection on Streams," Proceedings of The 33rd International Conference on Machine Learning 48 (June 2016): 2712–21.
[2] Bartos, Matthew D., A. Mullapudi, and S. C. Troutman. "rrcf: Implementation of the Robust Random Cut Forest Algorithm for Anomaly Detection on Streams." Journal of Open Source Software 4, no. 35 (2019): 1336.
Version History
Introduced in R2023b