kfoldPredict
Predict responses for observations in cross-validated quantile regression model
Since R2025a
Syntax
Description
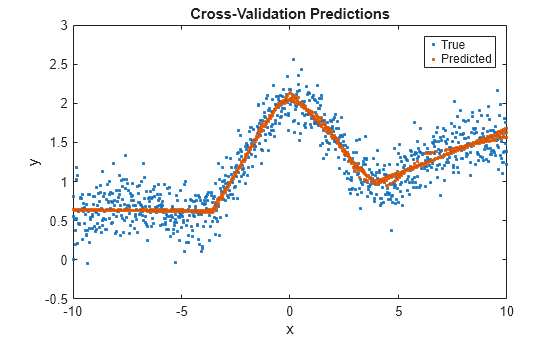
predictedY = kfoldPredict(CVMdl)CVMdl. For every fold, kfoldPredict predicts the
responses for validation-fold observations using a model trained on training-fold
observations. CVMdl.X and CVMdl.Y contain both sets of
observations.
predictedY = kfoldPredict(CVMdl,Name=Value)
[
additionally returns a matrix predictedY,crossingIndicator] = kfoldPredict(___)crossingIndicator whose entries indicate
whether predictions for the specified quantiles cross each other.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Algorithms
kfoldPredict computes predictions according to the
predict object function of the trained compact models in
CVMdl (CVMdl.Trained). For more information, see the
model-specific predict function reference pages in the following
table.
| Model Type | predict Function |
|---|---|
| Quantile linear regression model | predict |
| Quantile neural network model for regression | predict |
Version History
Introduced in R2025a