Deploy Signal Segmentation Deep Network on Raspberry Pi
This example details the workflow for waveform segmentation of an electrocardiogram (ECG) signal using short-time Fourier transform and a bidirectional long short-term memory (BiLSTM) network. The example also provides information on how to generate and deploy the code and the trained BiLSTM network for segmentation on a Raspberry Pi® target (ARM®-based device).
The pretrained network in the example is similar to the Waveform Segmentation Using Deep Learning example.
This example details:
Processor-in-the-loop (PIL) based workflow to verify generated code deployed and running on a Raspberry Pi from MATLAB®
Generation of a standalone executable
The PIL verification process is a crucial part of the design cycle to check that the behavior of the generated code matches the design before deploying a standalone executable.
ECG Dataset
This example uses ECG signals from the publicly available QT Database [1] [2]. The data consists of roughly 15 minutes of labeled ECG recordings, with a sample rate of 250 Hz, measured from a total of 105 patients.
The ECG signal can be divided into the following beat morphologies [3]:
P wave — A small deflection before the QRS complex representing atrial depolarization
QRS complex — Largest amplitude portion of the heartbeat
T wave — A small deflection after the QRS complex representing ventricular repolarization
The segmentation of these regions of ECG waveforms can provide the basis for measurements that assess the overall health of the human heart and the presence of abnormalities.
Prerequisites
ARM processor that supports the NEON extension
MATLAB Coder™
Embedded Coder®
Deep Learning Toolbox™
For supported versions of libraries and for information about setting up environment variables, see Prerequisites for Deep Learning with MATLAB Coder (MATLAB Coder).
Functionality of Generated Code
The core function in the generated executable:
Uses 15,000 samples of single-precision ECG data as input.
Computes the short-time Fourier transform of the signal.
Standardizes and normalizes the output.
Labels regions of the signal using the pretrained BiLSTM network.
Generates an output file with the labels.
The Entry-Point waveformSegmentation Function
An entry-point function, also known as the top-level or primary function, is a function you define for code generation. You must define an entry-point function that calls code-generation-enabled functions and generates C/C++ code from the entry-point function. All functions within the entry-point function must support code generation.
In this example, waveformSegmentation is the entry-point function. It takes an ECG signal as an input and passes it to the trained BiLSTM network for prediction. The performPreprocessing function preprocesses the raw signal and applies the short-time Fourier transform. The genClassifiedResults function passes the preprocessed signal to the network for prediction and displays the classification results.
type waveformSegmentationfunction out = waveformSegmentation(in)
%#codegen
% Copyright 2020-2024 The MathWorks, Inc.
persistent net;
if isempty(net)
net = coder.loadDeepLearningNetwork('trained-network-STFTBILSTM.mat', 'net');
end
preprocessedSignal = performPreprocessing(in);
out = cell(3,1);
% Specify coder.cstructname to lock down the structure name and ensure
% consistency with the one specified in the ecgsegmentation_main.cpp file.
coder.cstructname(out, 'waveformOutputs');
for indx = 1:3
dlIn = dlarray(preprocessedSignal{1,indx},"CT");
out{indx,1} = genClassifedResults(extractdata(net.predict(dlIn)));
end
end
Create a Connection to the Raspberry Pi
Use the MATLAB Support Package for Raspberry Pi function, raspi, to create a connection to the Raspberry Pi. In the following code, replace:
raspinamewith the host name of your Raspberry Piusernamewith your usernamepasswordwith your password
r = raspi(raspiname','username','password');
The example shows the PIL-based workflow for verification of code and design and then creates and deploys a standalone executable. Optionally, if you want to directly deploy a standalone executable, you can skip PIL execution and go to creating a standalone execution.
Generate PIL MEX Function
The first step shows a PIL-based workflow to generate a MEX function for the waveformSegmentation function.
Configure Code Generation Hardware Parameters for Raspberry Pi
Create a coder.EmbeddedCodeConfig object to generate a static library. Set the VerificationMode property to 'PIL' to enable PIL-based execution.
cfg = coder.config('lib','ecoder',true); cfg.VerificationMode = 'PIL';
Create a coder.Hardware object and specify the target hardware as Raspberry Pi. Assign hardware object to the Hardware property of cfg.
hw = coder.hardware('Raspberry Pi');
cfg.Hardware = hw;Create a deep learning configuration object and set target library to none. Attach the deep learning configuration object to the code generation configuration object.
dlcfg = coder.DeepLearningConfig(TargetLibrary = 'none');
cfg.DeepLearningConfig = dlcfg;Specify the build folder on the Raspberry Pi.
cfg.Hardware.BuildDir = '~/waveformSegmentation';Generate Source C++ Code Using codegen Function
Use the codegen function to generate the C++ code. When codegen is used with MATLAB Support Package for Raspberry Pi Hardware, the generated code is downloaded to the board and compiled there. A PIL MEX function is generated to communicate between MATLAB and the generated code running on the Raspberry Pi.
codegen -config cfg waveformSegmentation -args {coder.typeof(single(ones(1,15000)),[1,15000],[0,0])} -report
Run Executable Program on Raspberry Pi
Load the MAT file ecgsignal_test. The file stores a sample ECG signal on which you can test the generated code.
Run the generated waveformSegmentation_pil MEX function on the test signal.
load ecgsignal_test.mat;
out = waveformSegmentation_pil(test);### Starting application: 'codegen\lib\waveformSegmentation\pil\waveformSegmentation.elf'
To terminate execution: clear waveformSegmentation_pil
### Launching application waveformSegmentation.elf...
Display the signals with predicted labels.
labels = categorical(out{1}(1,2000:3000));
msk = signalMask(labels);
plotsigroi(msk,test(1,2000:3000))
title('Predicted Labels')
After the verifying the output of the PIL MEX function, you can create a standalone executable for the waveformSegmentation function.
The next part shows the code generation workflow to generate and deploy a standalone executable in the code for prediction on a Raspberry Pi using the MATLAB Coder App.
Create a Standalone Executable Using the MATLAB Coder App
The MATLAB Coder app generates C or C++ code from MATLAB code. The workflow-based user interface steps you through the code generation process. The following steps describe a brief workflow using the MATLAB Coder app. For more details, see MATLAB Coder (MATLAB Coder) and Generate C Code by Using the MATLAB Coder App (MATLAB Coder).
Select the Entry-Point Function File
On the MATLAB toolstrip Apps tab, under Code Generation, click the MATLAB Coder app icon. The app opens the Create MATLAB Coder Project dialog box. Provide the name of the project file and the folder in which you want to place the file. For this example, create a file named waveformSegmentation.coderprj in your current working folder.
A MATLAB Coder toolstrip opens that contains sections that correspond to the primary actions you perform when generating code, including preparing your MATLAB code for C/C++ code generation, generating code, performing verification on generated code, viewing the code generation report, and exporting the generated code.

In this state of the MATLAB Coder app, the Next Steps section of the code generation panel, add the entry-point function. Click the Add Entry Points button in the Inputs section of the code generation panel or the Entry Points button in the toolstrip. The Entry Points pane opens. Enter the name of your entry-point function waveformSegmentation.

Define Input Types
You must specify the properties of all entry-point function inputs. For this example, specify the value of the input in as single (1x15000). For more information on define input types in the MATLAB Coder App, refer to Generate C Code by Using the MATLAB Coder App (MATLAB Coder).

Generate Code
In the Prepare section of the MATLAB Coder toolstrip, make sure that Output is set to C and Build type is set to Executable (.exe). Click Settings button to open the standalone code generation settings dialog and:
In the Hardware pane, set Hardware Board to
Raspberry Piand enter the username and password parameters for the board.In the Custom Code pane, in additional source files, browse and select
ecgsegmentation_main.cpp. For more information on writing a C/C++ main function, refer to Structure of Generated Example C/C++ Main Function (MATLAB Coder).
Close the Settings window and click the Generate Code and Build button.
Fetch Generated Executable Directory
Once the code generation is complete, you can test the generated code on the Raspberry Pi. As a first step, copy the input ECG signal to the generated code directory. You can find the directory manually or by using the raspi.utils.getRemoteBuildDirectory API. This function lists the directories of the binary files that are generated by using the codegen function. Assuming that the binary is found in only one directory, enter:
applicationDirPaths = ...
raspi.utils.getRemoteBuildDirectory('applicationName','waveformSegmentation');
targetDirPath = applicationDirPaths{1}.directory;
Copy Input Files to the Raspberry Pi
To copy files required to run the executable program, use putFile, which is available with the MATLAB Support Package for Raspberry Pi Hardware. The input.csv file contains a sample ECG signal that is used to test the deployed code.
r.putFile('input.csv',targetDirPath);
input = dlmread('input.csv');
Run Executable Program on Raspberry Pi
Run the executable program on the Raspberry Pi from MATLAB and get the output file to MATLAB. Input file name should be passed as the command line argument for the executable.
exeName = 'waveformSegmentation.elf'; % Executable name
command = ['cd ' targetDirPath ';./' exeName];
system(r,command)
outputPath = strcat(targetDirPath,'/*.txt');
getFile(r,outputPath)
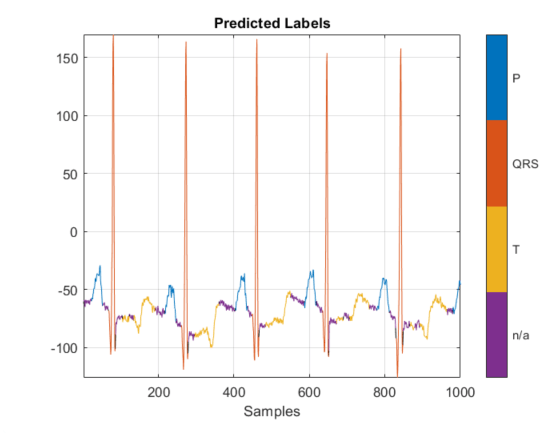
Display the signals with predicted labels. The output is depicted in the figure.
load ecgsignal_test.mat;
labels = categorical(textread('out.txt','%s')');
msk = signalMask(labels(1,2000:3000));
plotsigroi(msk,test(1,2000:3000))
title('Predicted Labels')
References
[1] McSharry, Patrick E., et al. "A dynamical model for generating synthetic electrocardiogram signals." IEEE® Transactions on Biomedical Engineering. Vol. 50, No. 3, 2003, pp. 289–294.
[2] Laguna, Pablo, Raimon Jané, and Pere Caminal. "Automatic detection of wave boundaries in multilead ECG signals: Validation with the CSE database." Computers and Biomedical Research. Vol. 27, No. 1, 1994, pp. 45–60.
[3] Goldberger, Ary L., Luis A. N. Amaral, Leon Glass, Jeffery M. Hausdorff, Plamen Ch. Ivanov, Roger G. Mark, Joseph E. Mietus, George B. Moody, Chung-Kang Peng, and H. Eugene Stanley. "PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals." Circulation. Vol. 101, No. 23, 2000, pp. e215–e220. [Circulation Electronic Pages; https://circ.ahajournals.org/content/101/23/e215.full].
See Also
Apps
- MATLAB Coder (MATLAB Coder)
Functions
codegen(MATLAB Coder) |fsst|signalMask
Topics
- Long Short-Term Memory Neural Networks (Deep Learning Toolbox)
- Generate C Code by Using the MATLAB Coder App (MATLAB Coder)
- Prerequisites for Deep Learning with MATLAB Coder (MATLAB Coder)
- Structure of Generated Example C/C++ Main Function (MATLAB Coder)