tcnAD
Create anomaly detector model that uses TCN network to detect anomalies
Since R2025a
Description
detector = tcnAD(numChannels)TcnDetector
model with numChannels channels for each time series input to the
detector.
detector = tcnAD( sets
additional options using one or more name-value arguments.numChannels,Name=Value)
For example, detector = tcnAD(3,DetectionWindowLength=20) creates a
detector model for data containing three input channels and with a detection window length
of 20.
Examples
Load the file sineWaveAnomalyData.mat, which contains two sets of synthetic three-channel sinusoidal signals.
sineWaveNormal contains 10 sinusoids of stable frequency and amplitude. Each signal has a series of small-amplitude impact-like imperfections. The signals have different lengths and initial phases.
load sineWaveAnomalyData.mat sineWaveNormal sineWaveAbnormal s1 = 3;
Plot input signals
Plot the first three normal signals. Each signal contains three input channels.
tiledlayout("vertical") ax = zeros(s1,1); for kj = 1:s1 ax(kj) = nexttile; plot(sineWaveNormal{kj}) title("Normal Signal Channels") end
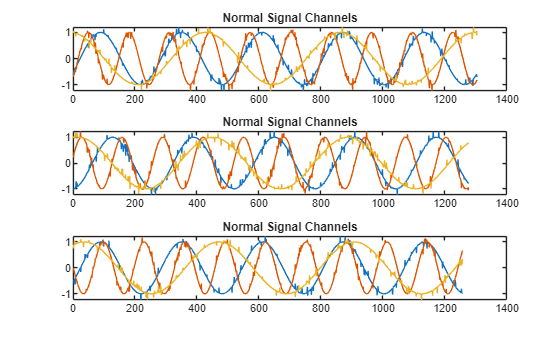
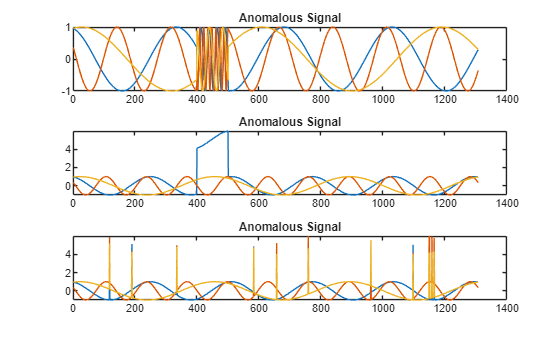
sineWaveAbnormal contains three signals, all of the same length. Each signal in the set has one or more anomalies.
All channels of the first signal have an abrupt change in frequency that lasts for a finite time.
The second signal has a finite-duration amplitude change in one of its channels.
The third signal has spikes at random times in all channels.
Plot the three signals with anomalies.
tiledlayout("vertical") ax = zeros(s1,1); for kj = 1:s1 ax(kj) = nexttile; plot(sineWaveAbnormal{kj}) title("Anomalous Signal") end
Create Detector
Use the tcnAD function to create a tcnDetector object with default options.
detector_tcn = tcnAD(3)
detector_tcn =
TcnDetector with properties:
FilterSize: 7
DropoutProbability: 0.2500
DetectionWindowLength: 10
DetectionStride: 10
NumFilters: 32
IsTrained: 0
NumChannels: 3
Layers: [23×1 nnet.cnn.layer.Layer]
Dlnet: [1×1 dlnetwork]
Threshold: []
ThresholdMethod: 'kSigma'
ThresholdParameter: 3
ThresholdFunction: []
Normalization: 'zscore'
Train Detector
Prepare to train detector_tcn by customizing a trainingOptions option set with a solver of "adam" and a limit of 100 for the number of training epochs.
trainopts = trainingOptions("adam",MaxEpochs=100);Train detector_tcn using the normal data and trainopts.
detector_tcn = train(detector_tcn,sineWaveNormal,trainingOpts=trainopts);
Iteration Epoch TimeElapsed LearnRate TrainingLoss
_________ _____ ___________ _________ ____________
1 1 00:00:00 0.001 1.6331
50 50 00:00:03 0.001 0.13097
100 100 00:00:06 0.001 0.085445
Training stopped: Max epochs completed
Computing threshold...
Threshold computation completed.
View the threshold that train computes and saves within detector_tcn. This computed value is influenced by random factors, such as which subsets of the data are used for training, and can change somewhat for different training sessions and different machines.
thresh = detector_tcn.Threshold
thresh = single
3.2729
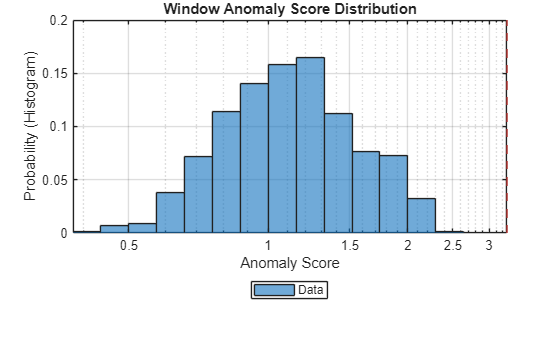
Plot the histogram of the anomaly scores for the normal data. Each score is calculated over a single detection window. The threshold, plotted as a vertical line, does not always completely bound the scores.
plotHistogram(detector_tcn,sineWaveNormal)
Use Detector to Identify Anomalies
Use the detect function to determine the anomaly scores for the anomalous data.
results = detect(detector_tcn, sineWaveAbnormal)
results=3×1 cell array
{130×3 table}
{130×3 table}
{130×3 table}
results is a cell array that contains three tables, one table for each channel. Each cell table contains three variables: WindowLabel, WindowAnomalyScore, and WindowStartIndices. Confirm the table variable names.
varnames = results{1}.Properties.VariableNamesvarnames = 1×3 cell
{'WindowLabels'} {'WindowAnomalyScores'} {'WindowStartIndices'}
Plot Results
Plot a histogram that shows the normal data, the anomalous data, and the threshold in one plot.
plotHistogram(detector_tcn,sineWaveNormal,sineWaveAbnormal)
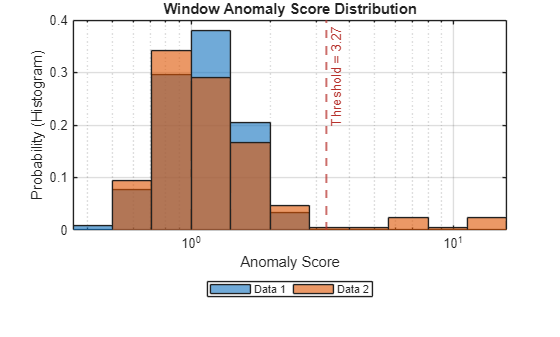
The histogram uses different colors for the normal and anomalous data.
Plot the detected anomalies of the third abnormal signal set.
plot(detector_tcn,sineWaveAbnormal{3})
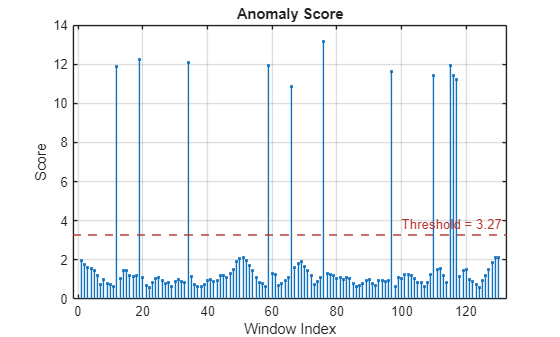
The top plot shows an overlay of red where the anomalies occur. The bottom plot shows how effective the threshold is at dividing the normal from the abnormal scores for Signal set 3.
Input Arguments
Number of input channels in each time series, specified as a positive integer. All time series inputs must have the same number of channels.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: detector = tcnAD(3,DetectionWindowLength=20) sets the length
of the detection window to 20.
Window
Window length of each time series segment, specified as a positive integer scalar.
Stride length of sliding window in detection
stage,
specified as a positive integer. DetectionStride controls the
number of overlapped samples. If you do not specify
DetectionStride, the software sets the stride length to the
value of detectionWindowLength to create non-overlapping
windows.
Threshold
Method to compute the detection threshold, specified as one of these values, each of which correspond to what the detection threshold is based on:
"kSigma"— Standard deviation of the normalized anomaly scores. The parameter k determines the threshold within the standard deviation levels that identifies an anomaly. The value of k is specified byThresholdParameter."contaminationFraction"— Percentage of anomalies within a specified fraction of windows, measured over the entire training set. The fraction value is specified byThresholdParameter."max"— Maximum window loss measured over the entire training data set and multiplied byThresholdParameter"mean"— Mean window loss measured over the entire training data set and multiplied byThresholdParameter"median"— Median window loss measured over the entire training data set and multiplied byThresholdParameter"manual"— Manual detection threshold value based onThreshold."customFunction"— Custom detection threshold method based onThresholdFunction.
If you specify ThresholdMethod, you can also specify
ThresholdParameter, Threshold, or ThresholdParameter. The
available threshold parameter depends on the specified detection method.
Anomaly score used to detect anomalies, specified as a positive scalar. The source
of the Threshold value depends on the setting of ThresholdMethod.
If
ThresholdMethodis"manual", you set the value.If
ThresholdMethodis"customFunction", the function specified inThresholdFunctioncomputes the value.For other values of
ThresholdMethod, useThresholdParameterto specify the detection threshold.
Parameter used for determining the detection threshold, specified as a numeric scalar.
The way you specify ThresholdParameter depends on the
specified value for ThresholdMethod. The following list describes
the specification of ThresholdParameter for each possible value
of ThresholdMethod
"kSigma"— SpecifyThresholdParameteras a positive numeric scalar. If you do not specifyThresholdParameter, the detector sets the threshold to 3."contaminationFraction"— SpecifyThresholdParameteras a as a nonnegative scalar less than 0.5. For example, if you specify"contaminationFraction"as0.05, then the threshold is set to identify the top 5% of the anomaly scores as anomalous. If you do not specifyThresholdParameter, the detector sets the threshold to 0.01."max","mean", or"median"— SpecifyThresholdParameteras a positive numeric scalar. If you do not specifyThresholdParameter, the detector sets the threshold to 1."customFunction"or"manual"—ThresholdParameterdoes not apply.
Function to compute custom detection threshold, specified as a function handle.
This argument applies only when ThresholdMethod is
specified as "customFunction".
The function must have two inputs
The first input is a vector of scalar window anomaly scores.
The second input is a vector representing all point-level anomalies.
For example, suppose that the value of
detectionWindowLengthis 10, the value ofdetectionStridesis set to be nonoverlapping, and the time series length is 10001. Then the first input vector has a length of 1000 and the second input vector has a length of 10000.The function must return a positive scalar corresponding to the detection threshold.
Model
Filter size of each convolutional layer, specified as a positive integer.
If you specify
FilterSizeas a scalar, the size of each filter is the same in all layers.
Number of filters in each convolutional layer, specified as a positive integer.
Dropout probability used to avoid overfitting, specified as a nonnegative numeric scalar less than 1. All convolution layers share the same dropout probability.
Normalization
Normalization technique for training and testing, specified as
"zscore", "range", or
"off".
Output Arguments
Anomaly detector model, returned as a TcnDetector object.
Version History
Introduced in R2025a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)