plotconfusion
Plot classification confusion matrix
Syntax
Description
plotconfusion(
plots a confusion matrix for the true labels targets,outputs)targets and
predicted labels outputs. Specify the labels as categorical
vectors, or in one-of-N (one-hot) form.
Tip
plotconfusion is not recommended for categorical
labels. Use confusionchart instead.
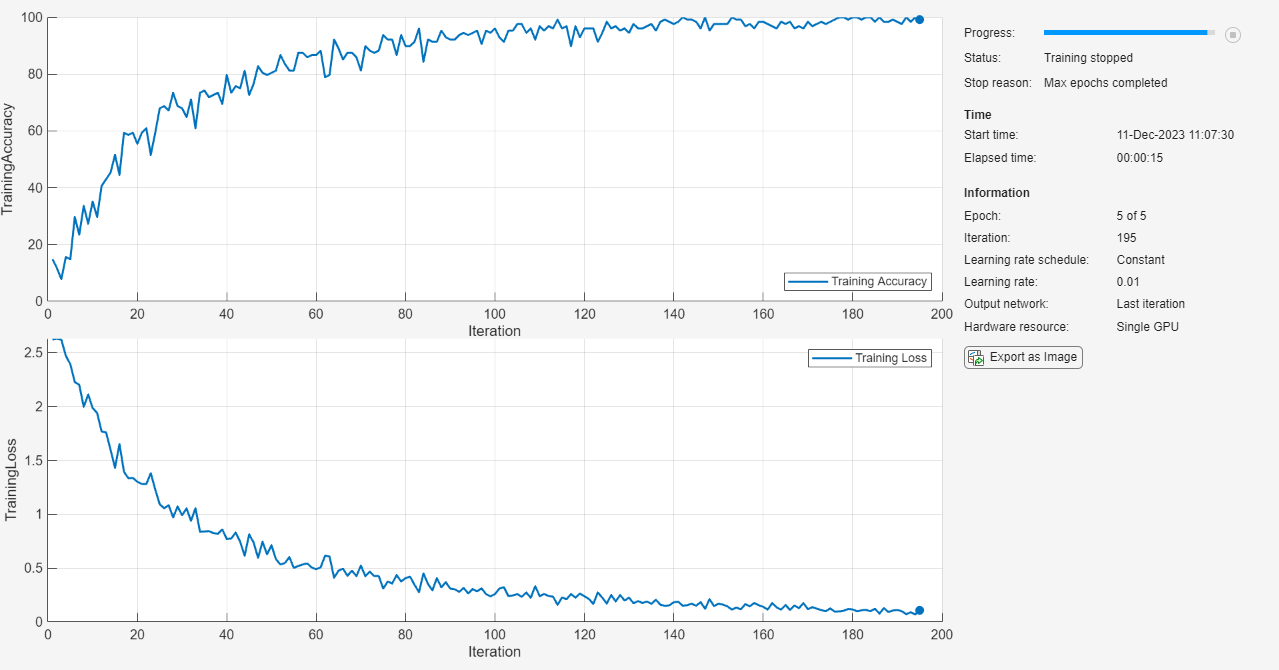
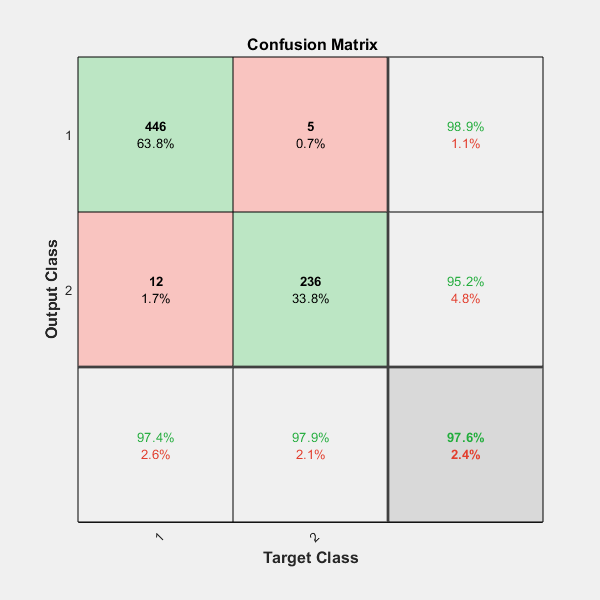
On the confusion matrix plot, the rows correspond to the predicted class (Output Class) and the columns correspond to the true class (Target Class). The diagonal cells correspond to observations that are correctly classified. The off-diagonal cells correspond to incorrectly classified observations. Both the number of observations and the percentage of the total number of observations are shown in each cell.
The column on the far right of the plot shows the percentages of all the examples predicted to belong to each class that are correctly and incorrectly classified. These metrics are often called the precision (or positive predictive value) and false discovery rate, respectively. The row at the bottom of the plot shows the percentages of all the examples belonging to each class that are correctly and incorrectly classified. These metrics are often called the recall (or true positive rate) and false negative rate, respectively. The cell in the bottom right of the plot shows the overall accuracy.
plotconfusion(targets1,outputs1,name1,targets2,outputs2,name2,...,targetsn,outputsn,namen)
plots multiple confusion matrices in one figure and adds the
name arguments to the beginnings of the titles of the
corresponding plots.
Examples
Input Arguments
Version History
Introduced in R2008a