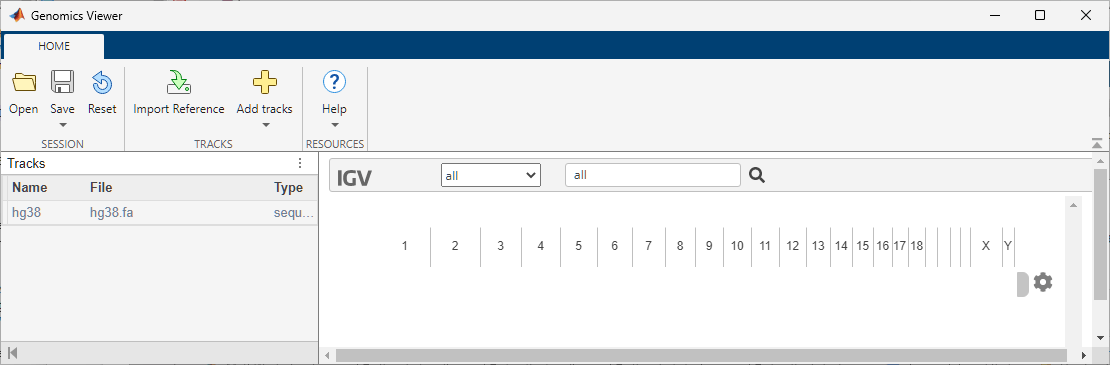
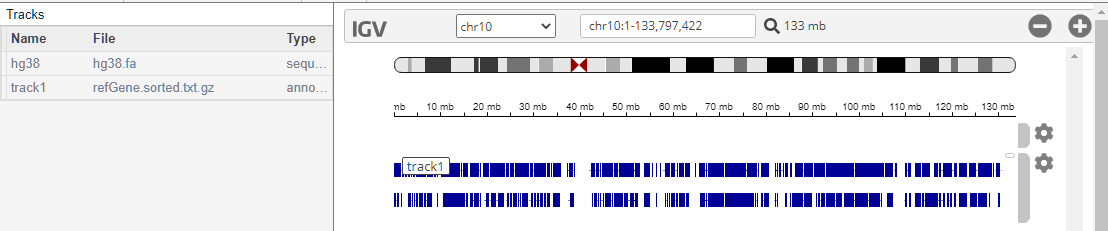
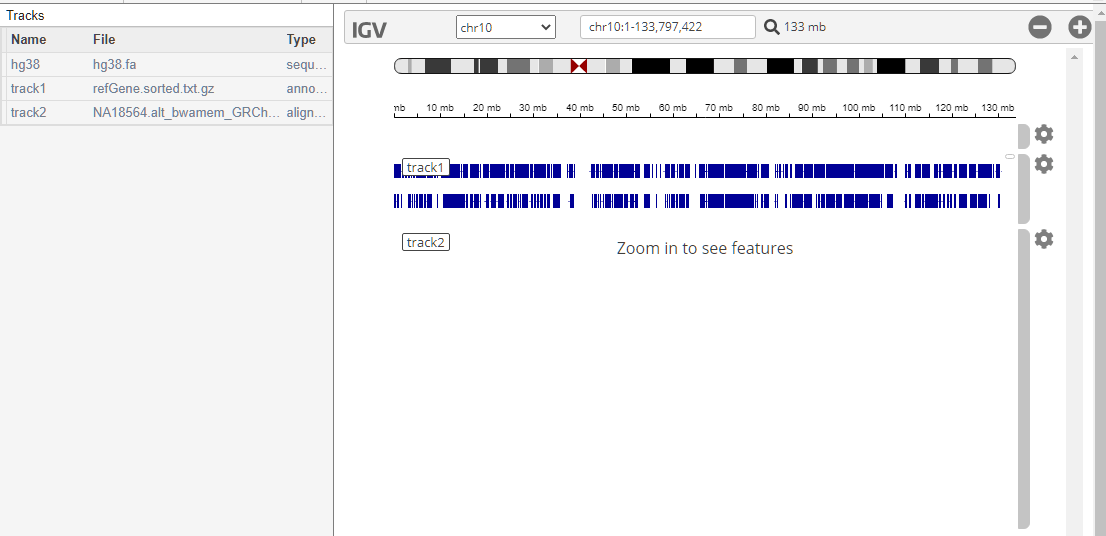
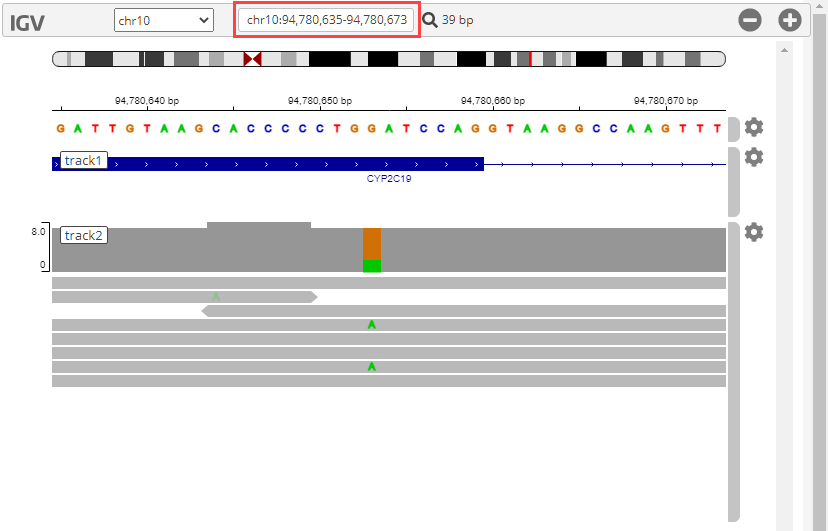
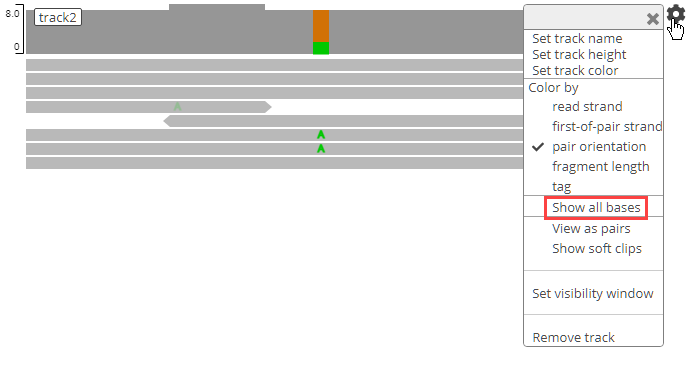
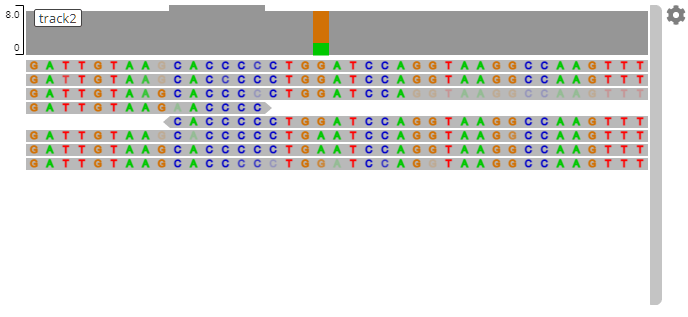
genomicsViewer
Description
Creation
Description
gv = genomicsViewergv, a
genomicsViewer object. When you call genomicsViewer
without specifying a reference sequence, the app uses Human
(GRCh38/hg38) as the reference sequence by default. If you are offline, you must
specify a local file to use as the reference by setting the ReferenceFile name-value
argument.
gv = genomicsViewer(Name=Value)ReferenceFile name-value argument to specify a reference
sequence.
Name-Value Arguments
Object Functions
addTracks | Add tracks to Genomics Viewer |
setReference | Specify reference sequence for Genomics Viewer |
close | Close Genomics Viewer app |
Examples
References
[1] Robinson, J., H. Thorvaldsdóttir, W. Winckler, M. Guttman, E. Lander, G. Getz, J. Mesirov. 2011. Integrative Genomics Viewer. Nature Biotechnology. 29:24–26.
[2] Thorvaldsdóttir, H., J. Robinson, J. Mesirov. 2013. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Briefings in Bioinformatics. 14:178–192.
Version History
Introduced in R2024a