The Particular Nature of Molecular Information in Engineered Biochemical Communication Systems
From the series: MathWorks Research Summit
Prof. Massimiliano Pierobon, Department of Computer Science and Engineering, University of Nebraska-Lincoln
Define and characterize information flow in communication systems based on molecule exchange and biochemical reactions, with the help of simulations realized through SimBiology®. The Molecular and Biochemical Telecommunications (MBiTe) Lab at the University of Nebraska-Lincoln, led by Dr. Massimiliano Pierobon, investigates communication systems based on the encoding, propagation, and decoding of information through biochemical reactions and molecule exchange. The definition of these systems ranges from molecule diffusion to the complex information flow at the basis of biological cells’ activity and multicellular organisms’ homeostasis. The biocompatibility and nanoscale feasibility make molecular communication a promising paradigm for engineering the interconnections between embedded computing systems able to not only interact with biological processes but also use these same processes as their building blocks, i.e., the Internet of Bio-Nano Things. The latter, enabled by emerging tools from synthetic biology for the genetic programming of biological cells’ behavior, opens novel application scenarios for communication and information theory, ranging from health care to environmental protection, defense, and consumer industries, towards the rational forward engineering of bio-sensing, stimulation, or actuation of biochemical processes, or even augmentation of the functionalities of the human body.
- Review the features of molecular communication theory as a paradigm, and its scope within the research in communication systems engineering.
- Survey the types of communication systems that have been realized through synthetic biology.
- Characterize a molecular communication system based on engineered cells with the development of an information theoretical framework to quantify biochemical information flow.
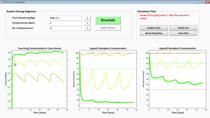
- Use SimBiology to simulate these engineered biochemical systems, and evaluate performance based on the results.
- Devise a novel methodology to engineer these systems with the synthetic biology formalism of biological circuits. This methodology is inspired by recent studies favoring the efficiency of analog computation over digital in biological cells, and it resulted in an award at the IEEE INFOCOM conference.
- By stemming from this methodology, review the design of a parity-check coding scheme implemented entirely in the biochemical domain, through activation and repression of genes, and biochemical reactions, rather than classical electrical circuits.
- Compare analytical formulations of information flow within this engineered coding scheme with simulation results based on SimBiology.
This talk concludes with the impact this research and the use of SimBiology had on the success of our international Genetically Engineered Machines (iGEM) team of undergraduate students.
This work was supported by the U.S. National Science Foundation under Grant CISE CCF-1816969.
Published: 28 Mar 2019