Export Model from Classification Learner to Experiment Manager
After training a classification model in Classification Learner, you can export the model to Experiment Manager to perform multiple experiments. By default, Experiment Manager uses Bayesian optimization to tune the model in a process similar to training optimizable models in Classification Learner. (For more information, see Hyperparameter Optimization in Classification Learner App.) Consider exporting a model to Experiment Manager when you want to do any of the following:
Adjust hyperparameter search ranges during hyperparameter tuning.
Change the training data.
Adjust the preprocessing steps that precede model fitting.
Tune hyperparameters using a different metric.
For a workflow example, see Tune Classification Model Using Experiment Manager.
Note that if you have a Statistics and Machine Learning Toolbox™ license, you do not need a Deep Learning Toolbox™ license to use the Experiment Manager app.
Export Classification Model
To create an Experiment Manager experiment from a model trained in Classification Learner, select the model in the Models pane. On the Learn tab, in the Export section, click Export Model and select Create Experiment.
Note
This option is not supported for binary GLM logistic regression models.
In the Create Experiment dialog box, modify the filenames or accept the default values.
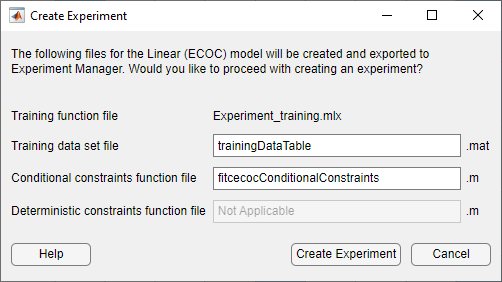
The app exports the following files to Experiment Manager:
Training function — This function trains a classification model using the model hyperparameters specified in the Experiment Manager app and records the resulting metrics and visualizations. For each trial of the experiment, the app calls the training function with a new combination of hyperparameter values, selected from the hyperparameter search ranges specified in the app. The app saves the returned trained model, which you can export to the MATLAB® workspace after training is complete.
Training data set — This
.matfile contains the full data set used in Classification Learner (including training and validation data, but excluding test data). Depending on how you imported the data into Classification Learner, the data set is contained in either a table nameddataTableor two separate variables namedpredictorMatrixandresponseData.Conditional constraints function — For some models, a conditional constraints function is required to tune model hyperparameters using Bayesian optimization. Conditional constraints enforce one of these conditions:
When some hyperparameters have certain values, other hyperparameters are set to given values.
When some hyperparameters have certain values, other hyperparameters are set to
NaNvalues (for numeric hyperparameters) or<undefined>values (for categorical hyperparameters).
For more information, see Conditional Constraints — ConditionalVariableFcn.
Deterministic constraints function — For some models, a deterministic constraints function is required to tune model hyperparameters using Bayesian optimization. A deterministic constraints function returns a
truevalue when a point in the hyperparameter search space is feasible (that is, the problem is valid or well defined at this point) and afalsevalue otherwise. For more information, see Deterministic Constraints — XConstraintFcn.
After you click Create Experiment, the app opens Experiment Manager. The Experiment Manager app then opens a dialog box in which you can choose to use a new or existing project for your experiment.

Select Hyperparameters
In Experiment Manager, use different hyperparameters and hyperparameter search ranges to tune your model. On the tab for your experiment, in the Hyperparameters section, click Add to add a hyperparameter to the model tuning process. In the table, double-click an entry to adjust its value.
You can also click the arrow next to Add and select Add From Suggested List to add hyperparameters that correspond to the exported model type.
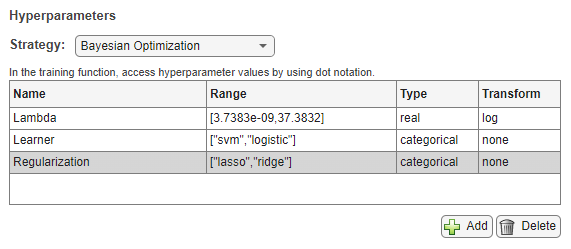
When you use the default Bayesian optimization strategy for model tuning, specify these properties of the hyperparameters used in the experiment:
Name — Enter a valid hyperparameter name.
Range — For a real- or integer-valued hyperparameter, enter a two-element vector that gives the lower bound and upper bound of the hyperparameter. For a categorical hyperparameter, enter an array of strings or a cell array of character vectors that lists the possible values of the hyperparameter.
Type — Select
realfor a real-valued hyperparameter,integerfor an integer-valued hyperparameter, orcategoricalfor a categorical hyperparameter.Transform — Select
noneto use no transform orlogto use a logarithmic transform. When you selectlog, the hyperparameter values must be positive. With this setting, the Bayesian optimization algorithm models the hyperparameter on a logarithmic scale.
The following table provides information about the hyperparameters you can tune in the app for each model type. The Suggested Hyperparameters column contains the hyperparameters that you can select in the Add From Suggested List dialog box.
| Model Type | Fitting Function | Default Hyperparameters | Suggested Hyperparameters |
|---|---|---|---|
| Tree | fitctree | For more
information, see | |
| Discriminant | fitcdiscr | For more
information, see | |
| Naive Bayes | fitcnb | For more information, see
| |
| SVM | fitcsvm (for two
classes), fitcecoc (for three
or more classes) |
|
For more
information, see |
| Efficient Linear | fitclinear (for two
classes), fitcecoc (for three
or more classes) | For more
information, see | |
| KNN | fitcknn | For more information, see | |
| Kernel | fitckernel (for two classes), fitcecoc (for three
or more classes) |
|
For more
information, see |
| Ensemble | fitcensemble |
For more
information, see | |
| Neural Network | fitcnet |
For more
information, see |
In the MATLAB Command Window, you can use the hyperparameters function to get more information about the
hyperparameters available for your model and their default search ranges. Specify
the fitting function, the training predictor data, and the training response
variable in the call to the hyperparameters function.
(Optional) Customize Experiment
In your experiment, you can change more than the model hyperparameters. In most cases, experiment customization requires editing the training function file before running the experiment. For example, to change the training data set, preprocessing steps, returned metrics, or generated visualizations, you must update the training function file. To edit the training function, click Edit in the Training Function section on the experiment tab. For an example that includes experiment customization, see Tune Classification Model Using Experiment Manager.
Some experiment customization steps do not require editing the training function file. For example, you can change the strategy for model tuning, adjust the Bayesian optimization options, or change the metric used to perform Bayesian optimization.
Change Strategy for Model Tuning
Instead of using Bayesian optimization to search for the best hyperparameter
values, you can sweep through a range of hyperparameter values. On the tab for
your experiment, in the Hyperparameters section, set
Strategy to Exhaustive
Sweep. In the hyperparameter table, enter the names and values of
the hyperparameters to use in the experiment. Hyperparameter values must be
scalars or vectors with numeric, logical, or string values, or cell arrays of
character vectors. For example, these are valid hyperparameter specifications,
depending on your model:
0.010.01:0.01:0.05[0.01 0.02 0.04 0.08]["Bag","AdaBoostM2","RUSBoost"]{'gaussian','linear','polynomial'}
When you run the experiment, Experiment Manager trains a model using every combination of the hyperparameter values specified in the table.
Adjust Bayesian Optimization Options
When you use Bayesian optimization, you can specify the duration of your
experiment. On the tab for your experiment, in the
Hyperparameters section, ensure that
Strategy is set to Bayesian
Optimization. In the Bayesian Optimization
Options section, enter the maximum time in seconds and the
maximum number of trials to run. Note that the actual run time and number of
trials in your experiment can exceed these settings because Experiment Manager
checks these options only when a trial finishes executing.
You can also specify the acquisition function for the Bayesian
optimization algorithm. In the Bayesian Optimization
Options section, click Advanced
Options. Select an acquisition function from the
Acquisition Function Name list. The default value for
this option is expected-improvement-plus. For more
information, see Acquisition Function Types.
Note that if you edit the training function file so that a new deterministic constraints function or conditional constraints function is required, you can specify the new function names in the Advanced Options section.
Change Metric Used to Perform Bayesian Optimization
By default, the app uses Bayesian optimization to try to find the combination
of hyperparameter values that minimizes the validation accuracy. You can specify
to minimize the validation total cost instead. On the tab for your experiment,
in the Metrics section, specify to optimize the
ValidationTotalCost value. Keep the
Direction set to
Minimize.
Note that if you edit the training function file to return another metric, you can specify it in the Metrics section. Ensure that the Direction is appropriate for the given metric.
Run Experiment
When are you ready to run your experiment, you can run it either sequentially or in parallel.
If you have Parallel Computing Toolbox™, Experiment Manager can perform computations in parallel. On the Experiment Manager tab, in the Execution section, select
Simultaneousfrom the Mode list.Note
Parallel computations with a thread pool are not supported in Experiment Manager.
Otherwise, use the default Mode option of
Sequential.
On the Experiment Manager tab, in the Run section, click Run.