Compare SimBiology Models
You can compare two SimBiology models and generate a list of differences between them. If the models contain diagram (graphical) information, the comparison results also include differences for the model diagrams. For details on how SimBiology compares and matches model components, see SimBiology Model Matching Policy. After comparing the models, you can also merge the differences. For details, see Merge SimBiology Models.
Compare Models Programmatically
Use sbiodiff at the command line. It returns a SimBiology.DiffResults object that contains the comparison
results.
Compare Models in Comparison Tool
To view the comparison results in the Comparison tool, do one of the following:
Command window — Call
visdiff(diffResults), where diffResults is theSimBiology.DiffResultsobject returned bysbiodiff.Alternatively, call
visdiffwith two SBPROJ files as inputs.Files panel — To compare two SBPROJ files on disk, select both files. Then, right-click and select Compare Selected Files/Folders. The Comparison tool opens the file you select first on the left.
Alternatively, right-click an SBPROJ file and select Compare To. Then, select the SBPROJ file you want to compare to.
The next figure shows an example of the Comparison tool comparing two models side by side. It uses different colors to indicate the insertion, deletion, and modification of model components. The tool lists the components in the order that they appear in the corresponding model.
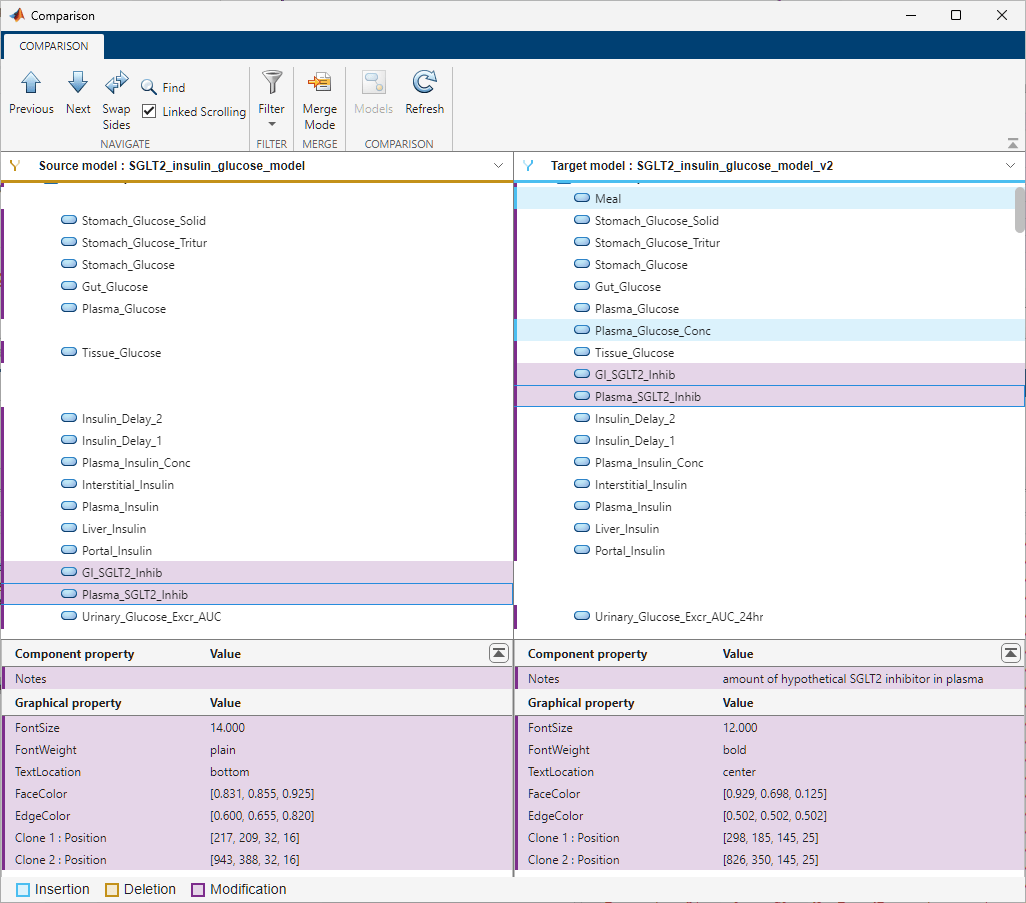
The Previous and Next buttons in the toolstrip let you step through each difference. Use the Swap Sides button to swap the Source model and Target model. Use the Models button to switch to different models if the input SBPROJ files contain multiple models. The Refresh button lets you refresh the comparison results for two models that are currently shown in the comparison tool. Use the Filter menu to customize the comparison results. The middle section of the app contains the tree of the matched model components. The bottom section shows the property values of the model components selected in the middle section.
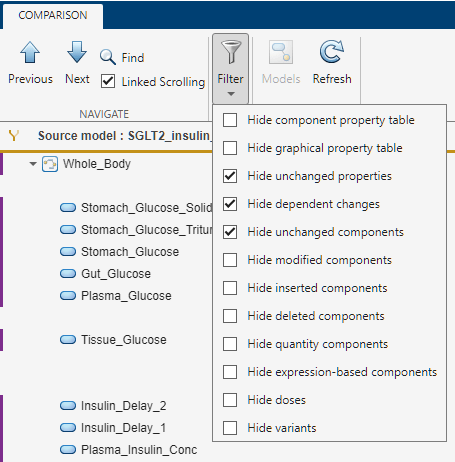
By default, the tool hides the unchanged properties, dependent changes, and unchanged components. The Filter menu gives additional criteria to fine-tune the results. For example, you can hide the quantity-based components (compartment, species, and parameters), the expression-based components (reactions, rules, events, and observables), and graphical changes.
Note
The dependent changes are the side effects of changing the
Name, Parent, or
Owner properties of a quantity component. When these
properties change, they trigger changes in other components referencing the
quantity component. For instance, the Reaction property of
a reaction C1.S1 -> C1.S2 changes to C1.S3 ->
C1.S2 when you rename the species S1 residing
in a compartment C1 to S3. The app hides
such dependent changes by default.
Note
The SimBiology comparison tool does not include annotation blocks in the comparison results.
Git Integration
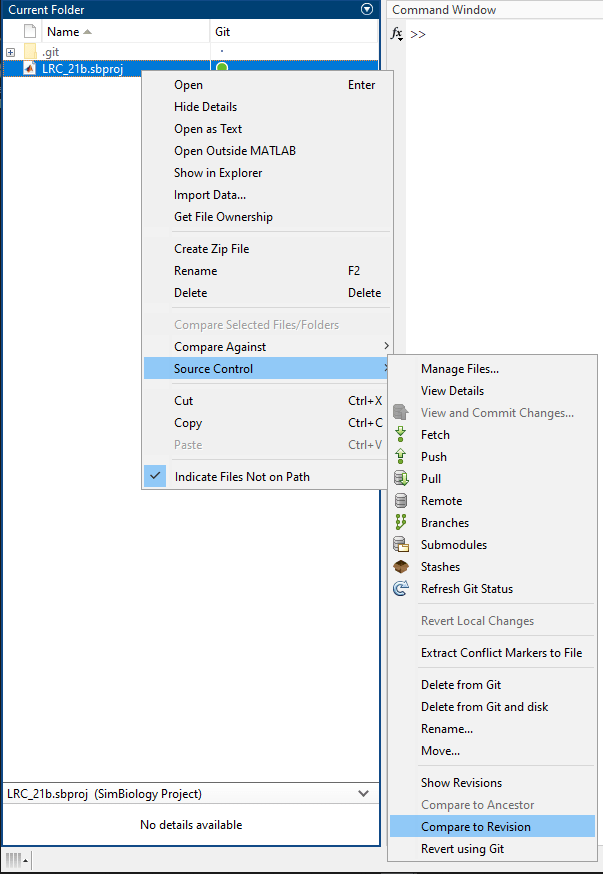
You can compare different versions of a SBPROJ file that you have in a Git repository from within MATLAB. Right-click the file and select Source Control > Show Revisions. In the Log dialog box, select the revision and click Compare to Local. See Compare Files and Folders and Merge Files for details. For instructions on setting up a Git repository, see Create Local Git Repository in MATLAB.
See Also
sbiodiff | SimBiology.DiffResults | getComponents | visdiff